| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,175,515 – 2,175,636 |
| Length | 121 |
| Max. P | 0.850773 |

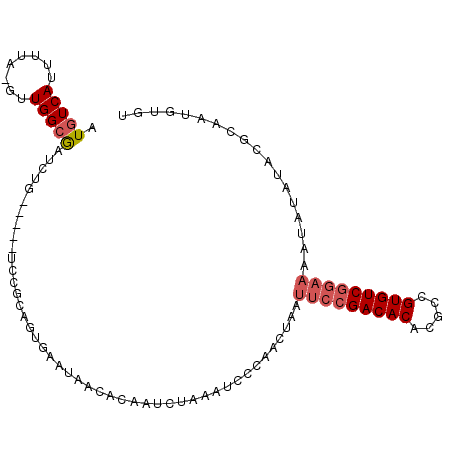
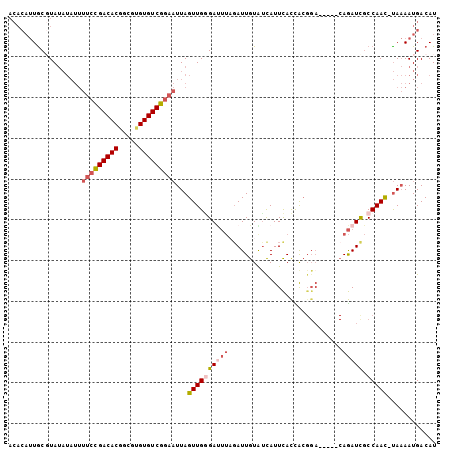
| Location | 2,175,515 – 2,175,609 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2175515 94 + 23771897 AUGUCAUUUUA-GUUGGCAAUCU----------CGGUGAAUAAUACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAGGUGU ........(((-(((((......----------...((.......)).........))))))))(((((((((.....)))))))))........(....).... ( -23.47) >DroPse_CAF1 266045 84 + 1 AUGUCAUUGUGUAUUGGGGAUUUGG----UCCGUAAUAAAUGACACAAUC----UCUCAACUAAAUCCGACACACGC-GUGUCAU--------UAAGCAGU---- .(((((((...(((((.((((...)----))).)))))))))))).....----..............(((((....-)))))..--------........---- ( -19.90) >DroSec_CAF1 227475 104 + 1 AUGUCAUUUUA-GUUGGCGAUCUGAUCUCUCCGCGGUGAAUAAUACAAUAUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGU ........(((-(((((.(((......((.(....).))(((......)))..)))))))))))(((((((((.....))))))))).((((((.....)))))) ( -24.60) >DroSim_CAF1 227908 99 + 1 AUGUCAUUUUA-GUUGGCGAUCUG-----UCCGCGGUGAAUAAUACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGU ........(((-(((((.((((((-----....))).................)))))))))))(((((((((.....))))))))).((((((.....)))))) ( -25.22) >DroEre_CAF1 232262 99 + 1 AUGUCAUUUUA-GUUGGCAAUCUG-----UCCGUAGUGAAUGACACAAUCUAAAUCGCAACUAAUUCCGACACGCGCCGUGUCGGAAAAUAUAUAAGCAAUGUGU .((((((((..-..((((.....)-----.)))....))))))))...................((((((((((...)))))))))).((((((.....)))))) ( -26.40) >DroYak_CAF1 225669 99 + 1 AUGUCAUUUUA-GUUGGCGAUCUG-----UCCGUAGUGAAUGGCACAAUCGAAGUCCCAACUAAUUCCGACACGCGCCGUGUCGGAAAAUAUAUACGUAAUGUGU ........(((-(((((((((.((-----(((((.....)))).))))))).....))))))))((((((((((...)))))))))).((((((.....)))))) ( -31.80) >consensus AUGUCAUUUUA_GUUGGCGAUCUG_____UCCGCAGUGAAUAACACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGU .(((((........))))).............................................(((((((((.....))))))))).................. (-15.35 = -15.97 + 0.61)

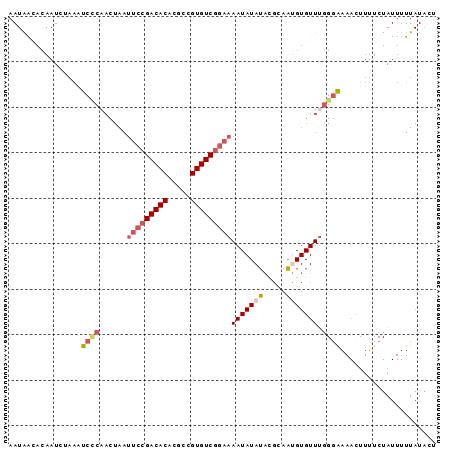
| Location | 2,175,515 – 2,175,609 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.72 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2175515 94 - 23771897 ACACCUUGCGUAUAUAUUUUCCGACACGGCGUGUGUCGGAAUUAGUUGGGAUUUAGAUUGUAUUAUUCACCG----------AGAUUGCCAAC-UAAAAUGACAU ......(((((.......(((((((((.....)))))))))(((((((((((((....((.......))...----------))))).)))))-))).))).)). ( -24.50) >DroPse_CAF1 266045 84 - 1 ----ACUGCUUA--------AUGACAC-GCGUGUGUCGGAUUUAGUUGAGA----GAUUGUGUCAUUUAUUACGGA----CCAAAUCCCCAAUACACAAUGACAU ----.((.((((--------(((((((-....))))).......)))))))----)...((((((((((((..(((----.....)))..))))...)))))))) ( -17.31) >DroSec_CAF1 227475 104 - 1 ACACAUUGCGUAUAUAUUUUCCGACACGGCGUGUGUCGGAAUUAGUUGGGAUUUAUAUUGUAUUAUUCACCGCGGAGAGAUCAGAUCGCCAAC-UAAAAUGACAU ...((((...........(((((((((.....)))))))))((((((((((((.....((.(((.(((......))).))))))))).)))))-))))))).... ( -28.40) >DroSim_CAF1 227908 99 - 1 ACACAUUGCGUAUAUAUUUUCCGACACGGCGUGUGUCGGAAUUAGUUGGGAUUUAGAUUGUAUUAUUCACCGCGGA-----CAGAUCGCCAAC-UAAAAUGACAU ...((((...........(((((((((.....)))))))))(((((((((((((.(.((((..........)))).-----)))))).)))))-))))))).... ( -29.00) >DroEre_CAF1 232262 99 - 1 ACACAUUGCUUAUAUAUUUUCCGACACGGCGCGUGUCGGAAUUAGUUGCGAUUUAGAUUGUGUCAUUCACUACGGA-----CAGAUUGCCAAC-UAAAAUGACAU ...((((...........((((((((((...))))))))))(((((((((((((.(.(((((........))))).-----))))))).))))-))))))).... ( -28.90) >DroYak_CAF1 225669 99 - 1 ACACAUUACGUAUAUAUUUUCCGACACGGCGCGUGUCGGAAUUAGUUGGGACUUCGAUUGUGCCAUUCACUACGGA-----CAGAUCGCCAAC-UAAAAUGACAU ...((((...........((((((((((...))))))))))((((((((.....(((((...((.........)).-----..))))))))))-))))))).... ( -31.00) >consensus ACACAUUGCGUAUAUAUUUUCCGACACGGCGUGUGUCGGAAUUAGUUGGGAUUUAGAUUGUAUCAUUCACCACGGA_____CAGAUCGCCAAC_UAAAAUGACAU ..................(((((((((.....)))))))))...((((((((((............................))))).)))))............ (-13.72 = -14.72 + 1.00)

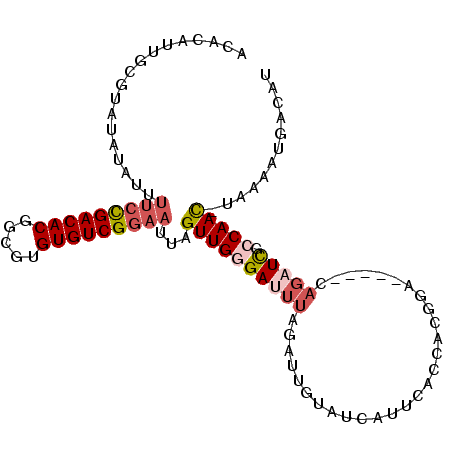
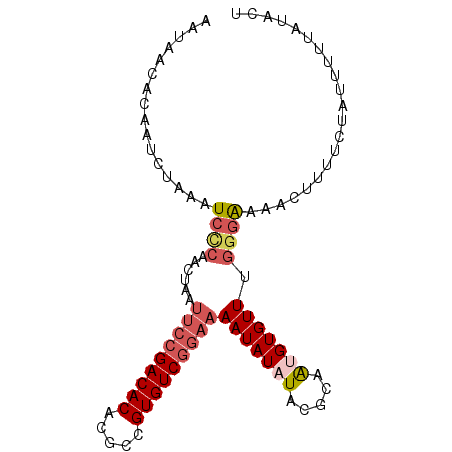
| Location | 2,175,542 – 2,175,636 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -14.59 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2175542 94 + 23771897 AAUAAUACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAGGUGUUGUGGGAAACUUUUCUAUUUUUAUACU ................(((((.....(((((((((.....)))))))))........(....)......))))).................... ( -23.40) >DroPse_CAF1 266079 72 + 1 AAUGACACAAUC----UCUCAACUAAAUCCGACACACGC-GUGUCAU--------UAAGCAGU------GGGAAAAGUUGUCUAUUUUUAU--- ...((((.....----(((((.........(((((....-)))))..--------.......)------)))).....)))).........--- ( -14.17) >DroSec_CAF1 227512 94 + 1 AAUAAUACAAUAUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGUUUGGGGAAACUUUUCUAUUUUUAUACU ................((((.......((((((((.....))))))))((((((((.....)))))))).)))).................... ( -23.20) >DroSim_CAF1 227940 93 + 1 AAUAAUACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGUUUGG-GAAACUUUUCUAUUUUUAUACU ................(((((.....(((((((((.....)))))))))(((((((.....))))))))))-)).................... ( -23.60) >DroEre_CAF1 232294 94 + 1 AAUGACACAAUCUAAAUCGCAACUAAUUCCGACACGCGCCGUGUCGGAAAAUAUAUAAGCAAUGUGUUUGGGAAAACUUUUCUAUUUUUAUAAU ................((.((.....((((((((((...))))))))))(((((((.....))))))))).))..................... ( -19.50) >DroYak_CAF1 225701 94 + 1 AAUGGCACAAUCGAAGUCCCAACUAAUUCCGACACGCGCCGUGUCGGAAAAUAUAUACGUAAUGUGUUUGGGAAAACUUUUCUAUUUUUAUAAU ................(((((.....((((((((((...))))))))))(((((((.....))))))))))))..................... ( -24.30) >consensus AAUAACACAAUCUAAAUCCCAACUAAUUCCGACACACGCCGUGUCGGAAAAUAUAUACGCAAUGUGUUUGGGAAAACUUUUCUAUUUUUAUACU ................((((......(((((((((.....)))))))))(((((((.....))))))).))))..................... (-14.59 = -15.80 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:23 2006