
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,242,348 – 19,242,502 |
| Length | 154 |
| Max. P | 0.963705 |

| Location | 19,242,348 – 19,242,462 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -6.15 |
| Energy contribution | -8.43 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19242348 114 + 23771897 UUUCAUUGUACACAAAGGUUUUGCAUUUGUUUGAGAAGACUGGCAAAAUAAAUGUACAU----AAAAGGAAAUGGUUUUCCAUAUUCACAGAGCACAUAUAGCA-CA-GAAUAUGUGCUC ......((((((.....(((((((....((((....))))..)))))))...)))))).----....(((.((((....)))).)))...((((((((((....-..-..)))))))))) ( -33.20) >DroSec_CAF1 70352 88 + 1 ----------------GGUUCUGCAUUUGUUCGAGUAGACUGGCAAAAUAAAUGUACAU----AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA-----------GAA-AUGUGCUU ----------------...(((((.(((((..(......)..)))))............----....(((.((((....)))).))))))))(((((-----------...-.))))).. ( -24.40) >DroSim_CAF1 75888 104 + 1 UUUCAUUGUACACAAAGGUUCUGCAUUUGUUCGAGUAGACGGGCAAAAUAAAUGUGCAU----AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA-----------GAA-AUGUGCUU ...................(((((.((((((((......))))))))............----....(((.((((....)))).))))))))(((((-----------...-.))))).. ( -29.10) >DroEre_CAF1 74850 119 + 1 UUUUAUUGUACACAAAGGAUCUGUAUGUGCUUGAGUAGACUAGGAAAAUAAAUGUAAAUGUAAUAAGGGACAUCCCUUUUCAUAUUCUCCUAGCGCACAUAGCAGCUACAA-AUGUGCUU .......(((((........((((((((((.......(.((((((.((((........((....(((((....)))))..)))))).))))))))))))).))))......-.))))).. ( -27.67) >DroYak_CAF1 101775 110 + 1 UUUUCUUGAACACAAAGGUUCUGCAUGUGCUUGAGUAGACUAGGAAAAUAAACAUACA-----AAAGGGCCAUCCAUUUCUAUUUCCCCAUAGU----AAGAUAGAUACAA-AUGUGUUU .......(((((((..((((((..((((.(((.((....))))).......))))...-----..)))))).......(((((((..(....).----.))))))).....-.))))))) ( -18.60) >consensus UUUCAUUGUACACAAAGGUUCUGCAUUUGUUUGAGUAGACUGGCAAAAUAAAUGUACAU____AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA_A____A____GAA_AUGUGCUU .....................(((((((((((.((....))....)))))))))))...........(((((....))))).........(((((((................))))))) ( -6.15 = -8.43 + 2.28)



| Location | 19,242,348 – 19,242,462 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -2.22 |
| Energy contribution | -5.18 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.09 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19242348 114 - 23771897 GAGCACAUAUUC-UG-UGCUAUAUGUGCUCUGUGAAUAUGGAAAACCAUUUCCUUUU----AUGUACAUUUAUUUUGCCAGUCUUCUCAAACAAAUGCAAAACCUUUGUGUACAAUGAAA ((((((((((..-..-....)))))))))).(((((...(((((....))))).)))----))((((((...((((((..((........))....)))))).....))))))....... ( -28.30) >DroSec_CAF1 70352 88 - 1 AAGCACAU-UUC-----------UGUGCUCUGCGAAUAUGGAAAACCAUUUCCUUUU----AUGUACAUUUAUUUUGCCAGUCUACUCGAACAAAUGCAGAACC---------------- ..(((((.-...-----------)))))((((((.....(((((....)))))....----............((((....((.....)).))))))))))...---------------- ( -16.60) >DroSim_CAF1 75888 104 - 1 AAGCACAU-UUC-----------UGUGCUCUGCGAAUAUGGAAAACCAUUUCCUUUU----AUGCACAUUUAUUUUGCCCGUCUACUCGAACAAAUGCAGAACCUUUGUGUACAAUGAAA .((((((.-...-----------))))))....(((.((((....))))))).....----.((((((....((((((...((.....))......))))))....))))))........ ( -20.20) >DroEre_CAF1 74850 119 - 1 AAGCACAU-UUGUAGCUGCUAUGUGCGCUAGGAGAAUAUGAAAAGGGAUGUCCCUUAUUACAUUUACAUUUAUUUUCCUAGUCUACUCAAGCACAUACAGAUCCUUUGUGUACAAUAAAA ..(((((.-......(((.((((((((((((((((((((((.(((((....))))).)))..........))))))))))))........))))))))))......)))))......... ( -36.52) >DroYak_CAF1 101775 110 - 1 AAACACAU-UUGUAUCUAUCUU----ACUAUGGGGAAAUAGAAAUGGAUGGCCCUUU-----UGUAUGUUUAUUUUCCUAGUCUACUCAAGCACAUGCAGAACCUUUGUGUUCAAGAAAA .((((((.-..((.(((.....----....((((((((((((.(..((.......))-----..)...))))))))))))..........((....)))))))...))))))........ ( -20.50) >consensus AAGCACAU_UUC____U____U_UGUGCUCUGCGAAUAUGGAAAACCAUUUCCUUUU____AUGUACAUUUAUUUUGCCAGUCUACUCAAACAAAUGCAGAACCUUUGUGUACAAUAAAA ..(((((................(((((...........(((((....)))))..........)))))....((((((..((........))....))))))....)))))......... ( -2.22 = -5.18 + 2.96)

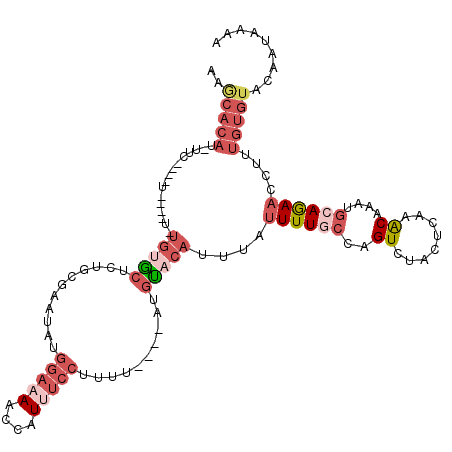
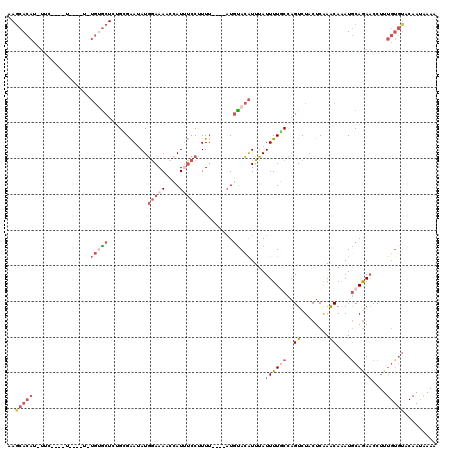
| Location | 19,242,388 – 19,242,502 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.82 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -6.57 |
| Energy contribution | -7.61 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.28 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19242388 114 + 23771897 UGGCAAAAUAAAUGUACAU----AAAAGGAAAUGGUUUUCCAUAUUCACAGAGCACAUAUAGCA-CA-GAAUAUGUGCUCUAAUGAACAGGAUAUUUGACCUAAUGUAAUUUGGUAAAAU .((..(((((..(((.(((----....(((.((((....)))).)))..(((((((((((....-..-..))))))))))).))).)))...)))))..))................... ( -28.30) >DroSec_CAF1 70376 104 + 1 UGGCAAAAUAAAUGUACAU----AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA-----------GAA-AUGUGCUUUAAUGAACAGAUUAUUCGAUCUAAUGUGAUUUUGUAAAAU ..(((((((....((.(((----....(((.((((....)))).)))..((((((((-----------...-.)))))))).))).))(((((....)))))......)))))))..... ( -26.20) >DroSim_CAF1 75928 104 + 1 GGGCAAAAUAAAUGUGCAU----AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA-----------GAA-AUGUGCUUUAACGAACAGGAUAUUUGAUCUAAUGUAAUUUUGAAAAAU ...((((((.....(((((----....(((((....)))))...((((.((((((((-----------...-.))))))))..))))..((((.....)))).)))))))))))...... ( -24.60) >DroEre_CAF1 74890 107 + 1 UAGGAAAAUAAAUGUAAAUGUAAUAAGGGACAUCCCUUUUCAUAUUCUCCUAGCGCACAUAGCAGCUACAA-AUGUGCUUCAACGAACUCGAUAUUCAAGCCUAUGUA------------ (((((.((((........((....(((((....)))))..)))))).)))))....((((((((((.((..-..)))))....((....))........).)))))).------------ ( -20.00) >DroYak_CAF1 101815 98 + 1 UAGGAAAAUAAACAUACA-----AAAGGGCCAUCCAUUUCUAUUUCCCCAUAGU----AAGAUAGAUACAA-AUGUGUUUUAAUGAACACGAAAUGCAACCCUAUGUA------------ ..............((((-----..((((.....((((((............((----(.......)))..-..((((((....))))))))))))...)))).))))------------ ( -16.60) >consensus UGGCAAAAUAAAUGUACAU____AAAAGGAAAUGGUUUUCCAUAUUCGCAGAGCACA_A____A____GAA_AUGUGCUUUAAUGAACAGGAUAUUCGACCUAAUGUAAUUU_G_AAAAU ..............(((((........(((((....)))))........((((((((................))))))))......................)))))............ ( -6.57 = -7.61 + 1.04)

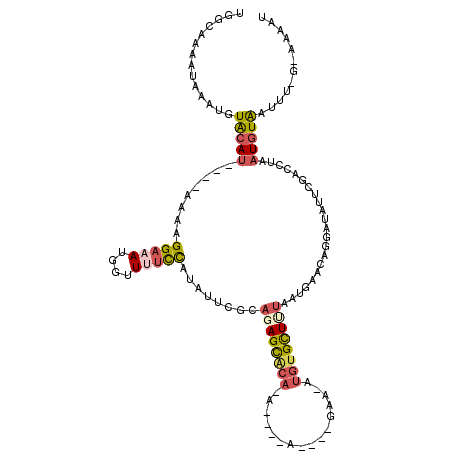
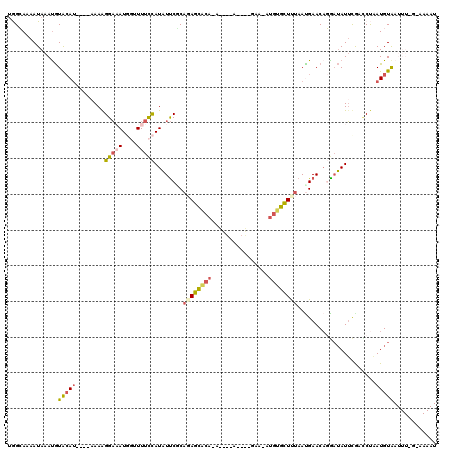
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:05 2006