| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,212,867 – 19,212,983 |
| Length | 116 |
| Max. P | 0.545094 |

| Location | 19,212,867 – 19,212,983 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -27.01 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19212867 116 - 23771897 UGCUCAUCAGCGGACAGGUUGCCAUCACGUCCGGAAGUCCC---UCUGUCCGGCUGUUUUGUGUCC-ACAUCUCAAUGGCAUCAAUAGCCACCGCUGAUUUCCUCUCUUUCUCUCCCUCU .....(((((((((((((..((..((......))..))..)---.))))))(((((((..(((.((-(........)))))).)))))))...))))))..................... ( -33.00) >DroSec_CAF1 41619 104 - 1 UGCUCAUCAGUGGACAGGUUGCCAUCACGUCCGGAAGUCCC---UCUGUCCGGCUGUUUUGUGUCC-ACAUCUCAAUGGCAUCAAUGGCCGCCGCUGAUUUCCUCUCU------------ .....((((((((...(((((((((.(((.(((((((....---.)).))))).)))..(((....-))).....))))))......))).)))))))).........------------ ( -31.10) >DroSim_CAF1 45358 112 - 1 UGCUCAUCAGCGGACAGGUUGCCAUCACGUCCGGAAGUCCC---UCUGUCCGGCUGUUUUGUGUCC-ACAUCUCAAUGGCAUCAAUGGCCGCCGCUGAUUUCCUCUCUCU----CCCUCU .....((((((((...(((((((((.(((.(((((((....---.)).))))).)))..(((....-))).....))))))......))).))))))))...........----...... ( -33.00) >DroEre_CAF1 44083 103 - 1 UGCUCAUCAGCGGACAGGUUGCCAUCCCGUCCGGAAGUCCC---UCUGUCCGGCUGUUUUGUGUCC-UCAACUCAAUGGCAUCAAUGGCCGCCGCUGAUUUCCUC-------------CU .....((((((((...(((((((((.(((..((((......---))))..))).((..(((.....-.)))..))))))))......))).))))))))......-------------.. ( -33.10) >DroYak_CAF1 70517 103 - 1 UGCUCAUCAGCGGACAGGUUGCCAUCACGUCCGGAAGUCCC---UCUGUCCGGCUGUUUUGUGUCC-ACAUCUCAAUGGCUUCAAUGGCCGCCGCUGAUUUCCUC-------------CU .....((((((((.(.((..((((..(((.(((((((....---.)).))))).)))..)).))))-..........((((.....)))))))))))))......-------------.. ( -32.70) >DroAna_CAF1 33222 114 - 1 UGCUCAUCAACGGACAGGUUGCCGCCACGGCCGGAAGUCCCUUCUCUGU-UGGCUGUUUUGUGUCCGACAUCUCAAUGGCAUCAAUGGCAGCCGCUGAUUUCCCGUCACU----CCGA-U ..........((((..((((((((.....((((((((...))))).(((-((((........).)))))).......))).....))))))))..((((.....)))).)----))).-. ( -34.10) >consensus UGCUCAUCAGCGGACAGGUUGCCAUCACGUCCGGAAGUCCC___UCUGUCCGGCUGUUUUGUGUCC_ACAUCUCAAUGGCAUCAAUGGCCGCCGCUGAUUUCCUCUCU__________CU .....((((((((...((((((((..(((.(((((((........)).))))).))).(((.(........).))))))))......))).))))))))..................... (-27.01 = -27.43 + 0.42)

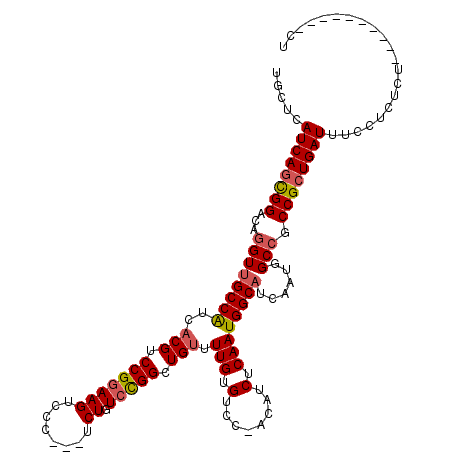
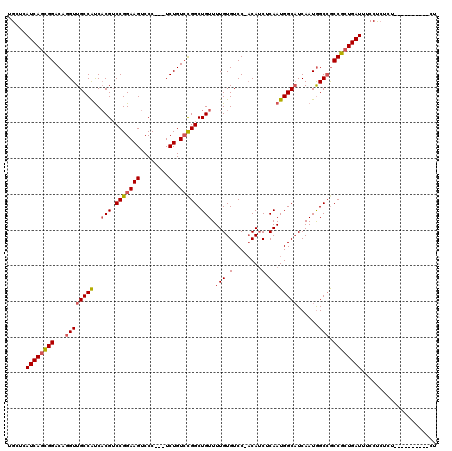
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:45 2006