
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,206,059 – 19,206,198 |
| Length | 139 |
| Max. P | 0.999801 |

| Location | 19,206,059 – 19,206,158 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -25.37 |
| Energy contribution | -25.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19206059 99 + 23771897 GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ..........((.((((((....((((((((((....(((((------------((......)))))))....)))))))).......((.....)))).)))))).)).. ( -31.90) >DroSec_CAF1 34407 99 + 1 GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ..........((.((((((....((((((((((....(((((------------((......)))))))....)))))))).......((.....)))).)))))).)).. ( -31.90) >DroSim_CAF1 38733 99 + 1 GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ..........((.((((((....((((((((((....(((((------------((......)))))))....)))))))).......((.....)))).)))))).)).. ( -31.90) >DroEre_CAF1 37096 99 + 1 GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGC------------GGAAAAUGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ..........((.((((((....((((((((((....(((((------------((......)))))))....)))))))).......((.....)))).)))))).)).. ( -31.90) >DroYak_CAF1 62079 111 + 1 GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGCGGAAAGUGCUGCGGAAAAUGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ......(((..(((.(((((((.(((.(((((((((((((......)))))).)))))))...)))..)))))))....(((((..........)))))....))).))). ( -36.90) >consensus GAAAGGCGUAGCACACUUUCGGGGCGUUUUUCCGGGCGCUGC____________GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGA ..........((.((((((....((((((((((....(((((......................)))))....)))))))).......((.....)))).)))))).)).. (-25.37 = -25.37 + -0.00)

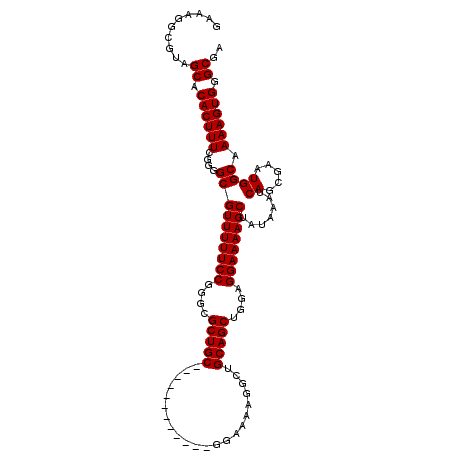
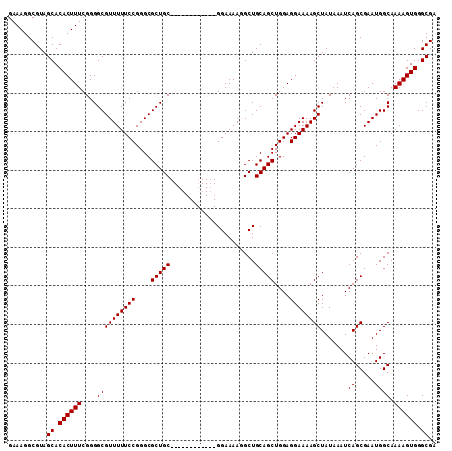
| Location | 19,206,059 – 19,206,158 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -25.87 |
| Energy contribution | -25.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19206059 99 - 23771897 UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....))))((((((....(((((..........------------)))))....))))))......)))))))).)).......... ( -27.20) >DroSec_CAF1 34407 99 - 1 UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....))))((((((....(((((..........------------)))))....))))))......)))))))).)).......... ( -27.20) >DroSim_CAF1 38733 99 - 1 UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....))))((((((....(((((..........------------)))))....))))))......)))))))).)).......... ( -27.20) >DroEre_CAF1 37096 99 - 1 UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCAUUUUCC------------GCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....))))((((((....(((((..........------------)))))....))))))......)))))))).)).......... ( -27.20) >DroYak_CAF1 62079 111 - 1 UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCAUUUUCCGCAGCACUUUCCGCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....(((((........))))))))).(((((((..((.((......)).)).)))))))......)))))))).)).......... ( -29.40) >consensus UCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC____________GCAGCGCCCGGAAAAACGCCCCGAAAGUGUGCUACGCCUUUC ..((.((((((((......((((.....))))((((((....(((((......................)))))....))))))......)))))))).)).......... (-25.87 = -25.87 + -0.00)

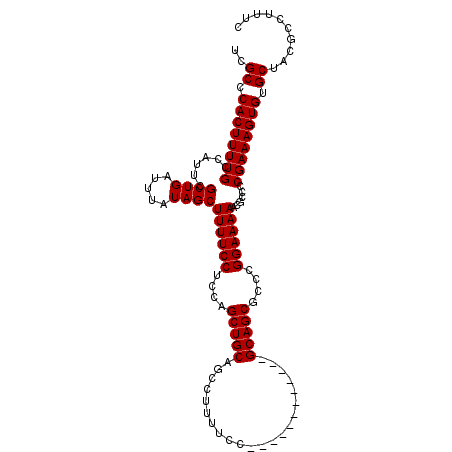
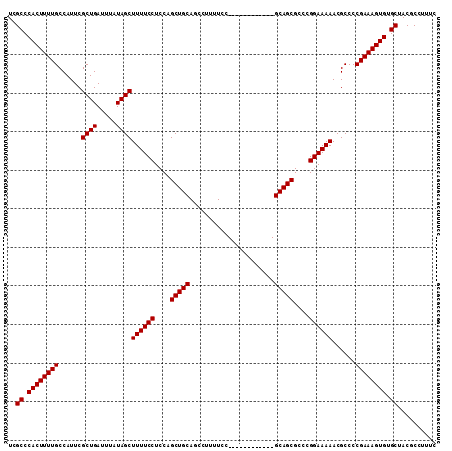
| Location | 19,206,099 – 19,206,198 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.06 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19206099 99 + 23771897 GC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCAGCCACCAUGGAUGCAGUUUUCCU ..------------(((((((((((((((..........(((((..........))))).((((((.(......).))))))))))))))).(....)......)))))). ( -31.90) >DroSec_CAF1 34447 99 + 1 GC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCAGCCACCGUGGAUGCAGUUUGCCA ..------------((...(((((((((((........))))....(((.(((..((((........(((....)))((......)).)))).))).)))))))))).)). ( -32.10) >DroSim_CAF1 38773 99 + 1 GC------------GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCAGCCACCGUGGAUGCAGUUUCCCU ((------------((....(((((((((..........(((((..........))))).((((((.(......).))))))))))))))).))))(((.......))).. ( -32.10) >DroEre_CAF1 37136 99 + 1 GC------------GGAAAAUGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCACCCACCAUGGAUGCAGUUUUCCG .(------------(((((..(((((((((........))))....(((.((.....))....(((((.(.(((((.....)))))).)))))....)))))))))))))) ( -32.90) >DroYak_CAF1 62119 111 + 1 GCGGAAAGUGCUGCGGAAAAUGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCACCCACCGUGGAUGCAGUUUUCCU ((((......))))(((((..(((((((((........))))....(((.((.....))....(((((.(.(((((.....)))))).)))))....))))))))))))). ( -36.30) >consensus GC____________GGAAAAGGCUGCAGCUGGAGGAAAAGCUAUAAAUCAGCGAAUGGCAAAAGUGGGCGACAACGCGCUUUGUUGCAGCCACCGUGGAUGCAGUUUUCCU ..............(((((..((((((.((((.((....(((.......))).....((((....(.(((....))).)....))))..)).))).)..))))))))))). (-28.90 = -29.06 + 0.16)



| Location | 19,206,099 – 19,206,198 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19206099 99 - 23771897 AGGAAAACUGCAUCCAUGGUGGCUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GC .(((((((((((...((((((((.(((((.......))))).))).......)))))..((((.................)))))))))..))))))------------.. ( -28.54) >DroSec_CAF1 34447 99 - 1 UGGCAAACUGCAUCCACGGUGGCUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GC ((((((((((......))))(((.(((((.......))))).))).....))))))...((((.....(((((........))))))))).......------------.. ( -26.70) >DroSim_CAF1 38773 99 - 1 AGGGAAACUGCAUCCACGGUGGCUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC------------GC .(((((((((((.....((((((.(((((.......))))).))).......)))....((((.................)))))))))..))))))------------.. ( -26.94) >DroEre_CAF1 37136 99 - 1 CGGAAAACUGCAUCCAUGGUGGGUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCAUUUUCC------------GC (((((((..(((.....((((((((((((.......)))).))))))))..))).....((((.....(((((........))))))))).))))))------------). ( -35.00) >DroYak_CAF1 62119 111 - 1 AGGAAAACUGCAUCCACGGUGGGUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCAUUUUCCGCAGCACUUUCCGC .(((((.((((......((((((((((((.......)))).))))))))..........((((.....(((((........))))))))).......))))...))))).. ( -36.40) >consensus AGGAAAACUGCAUCCACGGUGGCUGCAACAAAGCGCGUUGUCGCCCACUUUUGCCAUUCGCUGAUUUAUAGCUUUUCCUCCAGCUGCAGCCUUUUCC____________GC .((((((((((.....(((((..(((((......(((....)))......))).))..)))))......((((........))))))))..)))))).............. (-24.32 = -24.16 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:36 2006