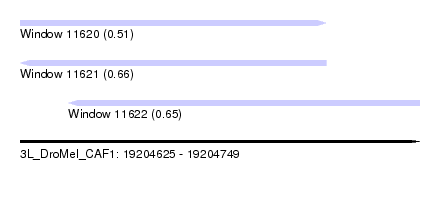
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,204,625 – 19,204,749 |
| Length | 124 |
| Max. P | 0.656153 |

| Location | 19,204,625 – 19,204,720 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -22.42 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
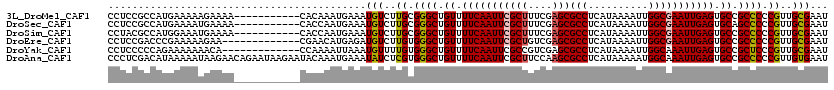
>3L_DroMel_CAF1 19204625 95 + 23771897 -------------------------CACUAUAUCUGUCUGAUUCGCAACGGGGGCGGCACUCAAUUCGCCAAUUUUAUGAGGCGCUCGAAAGCGAAUUGAAAACAGCCCGCAAGACAUUU -------------------------.........((((((.....)...(.((((.....((((((((((..........)))(((....)))))))))).....)))).).)))))... ( -26.90) >DroSec_CAF1 32982 95 + 1 -------------------------CACUAUAUCUGUCUGAUUCGCAACGGGGGCUGCACUCAAUUCGCCAAUUUUAUGAGGCGCUCGAAAGCGAAUUGAAAACAGCCCGCAAGACAUUU -------------------------.........((((((.....)...(.((((((...((((((((((..........)))(((....))))))))))...)))))).).)))))... ( -30.50) >DroSim_CAF1 37317 95 + 1 -------------------------CACUAUAUCUGUCUGAUUCGCAACGGGGGCGGCACUCAAUUCGCCAAUUUUAUGAGGCGCUCGAAAGCGAAUUGAAAACAGCCCGCAAGACAUUU -------------------------.........((((((.....)...(.((((.....((((((((((..........)))(((....)))))))))).....)))).).)))))... ( -26.90) >DroEre_CAF1 35567 113 + 1 -U-----CGUGCUGGGGUACGAGUUC-CUAUAUCUGUCUGAUUCGCAACGGGGGCGGCACUCAAUUCGCCAAUUUUAUGAGGCGCUCGACAGCGAAUUGAAAACAGCCCACAAGACAUCU -(-----(((((....))))))....-.......(((((...(((...)))((((.....((((((((((..........)))(((....)))))))))).....))))...)))))... ( -33.10) >DroYak_CAF1 60561 114 + 1 -U-----CGGGCUGGGGUACGAGUUCACUAUAUCUGUCUGAUUCGCAACGGGAGCGGCACUCAAUUUGCCAAUUUUAUGAGGCGCUCGACGGCGAAUUGAAAACAGCCCACAAAACAUUU -.-----.(((((((((((..((....)).))))).((.(((((((..((.(((((.(.((((..............))))))))))..)))))))))))...))))))........... ( -35.84) >DroAna_CAF1 23579 120 + 1 AUUAGGGAGGGCGGGGCUGGGGGCAUACUAUAUCUGUCCGAUUCACAACGGGGGCGGCACUCAAUUUGCCAUUUUUAUGAGGCGCUUGGAAGCGAAUUGAAAACAGCCCACGAGAUAUUU ...........((((((((.(((((.........)))))(((((.(..((((.((((((.......)))).(((....))))).))))...).))))).....)))))).))........ ( -34.00) >consensus _________________________CACUAUAUCUGUCUGAUUCGCAACGGGGGCGGCACUCAAUUCGCCAAUUUUAUGAGGCGCUCGAAAGCGAAUUGAAAACAGCCCACAAGACAUUU ..................................((((((..((.....))((((.....((((((((((..........)))(((....)))))))))).....)))).)).))))... (-22.42 = -21.98 + -0.44)

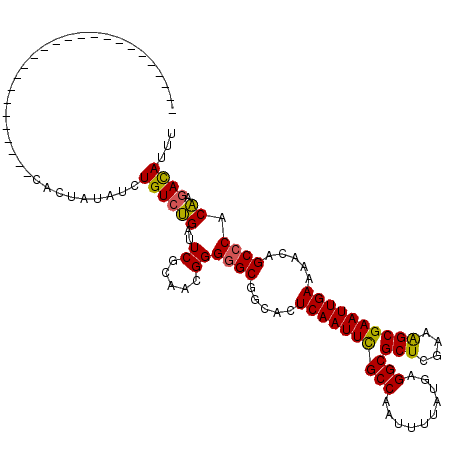
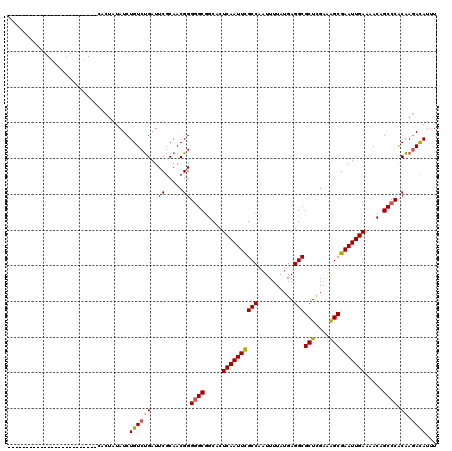
| Location | 19,204,625 – 19,204,720 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
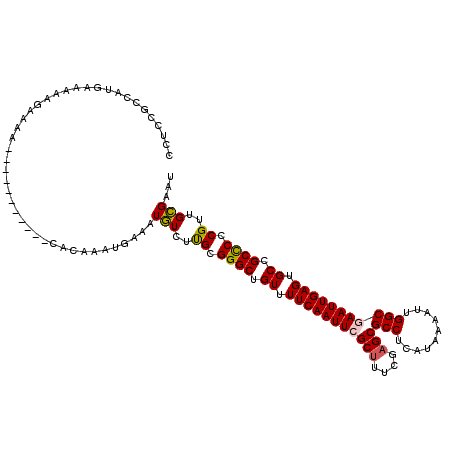
>3L_DroMel_CAF1 19204625 95 - 23771897 AAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAUCAGACAGAUAUAGUG------------------------- ..((((((.(.((((.((.(((((((((((....)))(((..........))))))))))).)).)))).)(((........)))))))))....------------------------- ( -30.50) >DroSec_CAF1 32982 95 - 1 AAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCAGCCCCCGUUGCGAAUCAGACAGAUAUAGUG------------------------- ..((((((.(.(((((((.(((((((((((....)))(((..........))))))))))).))))))).)(((........)))))))))....------------------------- ( -35.10) >DroSim_CAF1 37317 95 - 1 AAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAUCAGACAGAUAUAGUG------------------------- ..((((((.(.((((.((.(((((((((((....)))(((..........))))))))))).)).)))).)(((........)))))))))....------------------------- ( -30.50) >DroEre_CAF1 35567 113 - 1 AGAUGUCUUGUGGGCUGUUUUCAAUUCGCUGUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAUCAGACAGAUAUAG-GAACUCGUACCCCAGCACG-----A- ..((((((.(.((((.((.(((((((((((....)))(((..........))))))))))).)).)))).)(((........)))))))))..-....((((........)))-----)- ( -32.10) >DroYak_CAF1 60561 114 - 1 AAAUGUUUUGUGGGCUGUUUUCAAUUCGCCGUCGAGCGCCUCAUAAAAUUGGCAAAUUGAGUGCCGCUCCCGUUGCGAAUCAGACAGAUAUAGUGAACUCGUACCCCAGCCCG-----A- ..........(((((((......((((((((..((((((((((..............)))).).))))).))..))))))........(((((....)).)))...)))))))-----.- ( -30.54) >DroAna_CAF1 23579 120 - 1 AAAUAUCUCGUGGGCUGUUUUCAAUUCGCUUCCAAGCGCCUCAUAAAAAUGGCAAAUUGAGUGCCGCCCCCGUUGUGAAUCGGACAGAUAUAGUAUGCCCCCAGCCCCGCCCUCCCUAAU ..((((((.((((((.((.(((((((.(((....)))(((..........))).))))))).)).))))(((........)))))))))))............................. ( -28.90) >consensus AAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAUCAGACAGAUAUAGUG_________________________ ..((((((.(.((((.((.(((((((((((....)))(((..........))))))))))).)).)))).)(((........)))))))))............................. (-27.40 = -27.48 + 0.09)

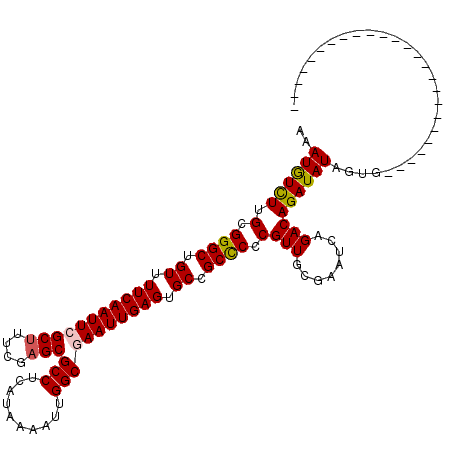
| Location | 19,204,640 – 19,204,749 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19204640 109 - 23771897 CCUCCGCCAUGAAAAAGAAAA-----------CACAAAUGAAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAU ....(((.(((...((((...-----------..(....).....))))..((((.((.(((((((((((....)))(((..........))))))))))).)).)))).))).)))... ( -31.60) >DroSec_CAF1 32997 109 - 1 CCUCCGCCAUGAAAAUGAAAA-----------CACCAAUGAAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCAGCCCCCGUUGCGAAU ....((((((....)))....-----------.................(.(((((((.(((((((((((....)))(((..........))))))))))).))))))).)...)))... ( -35.90) >DroSim_CAF1 37332 109 - 1 CCUACGCCAUGGAAAUGAAAA-----------CACCAAUGAAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAU ....(((.((((.........-----------...(((.((....))))).((((.((.(((((((((((....)))(((..........))))))))))).)).)))))))).)))... ( -34.20) >DroEre_CAF1 35600 107 - 1 CCUCCGACCCGAAAAAGAA-------------CGAACAUGAGAUGUCUUGUGGGCUGUUUUCAAUUCGCUGUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAU ..((((((..........(-------------(((((((...)))).))))((((.((.(((((((((((....)))(((..........))))))))))).)).))))..)))).)).. ( -31.20) >DroYak_CAF1 60595 107 - 1 CCUCCCCCAGAAAAAAACA-------------CCAAAAUUAAAUGUUUUGUGGGCUGUUUUCAAUUCGCCGUCGAGCGCCUCAUAAAAUUGGCAAAUUGAGUGCCGCUCCCGUUGCGAAU .........(((((...((-------------(.(((((.....)))))))).....))))).((((((((..((((((((((..............)))).).))))).))..)))))) ( -25.04) >DroAna_CAF1 23619 120 - 1 CCCUCGACAUAAAAAUAAGAACAGAAUAAGAAUACAAAUGAAAUAUCUCGUGGGCUGUUUUCAAUUCGCUUCCAAGCGCCUCAUAAAAAUGGCAAAUUGAGUGCCGCCCCCGUUGUGAAU ..(.((((.............................((((......))))((((.((.(((((((.(((....)))(((..........))).))))))).)).))))..)))).)... ( -25.70) >consensus CCUCCGCCAUGAAAAAGAAAA___________CACAAAUGAAAUGUCUUGCGGGCUGUUUUCAAUUCGCUUUCGAGCGCCUCAUAAAAUUGGCGAAUUGAGUGCCGCCCCCGUUGCGAAU ...........................................(((..((.((((.((.(((((((((((....)))(((..........))))))))))).)).)))).))..)))... (-26.40 = -26.35 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:32 2006