
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,198,093 – 19,198,331 |
| Length | 238 |
| Max. P | 0.999912 |

| Location | 19,198,093 – 19,198,213 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -39.17 |
| Energy contribution | -39.03 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999912 |
| Prediction | RNA |
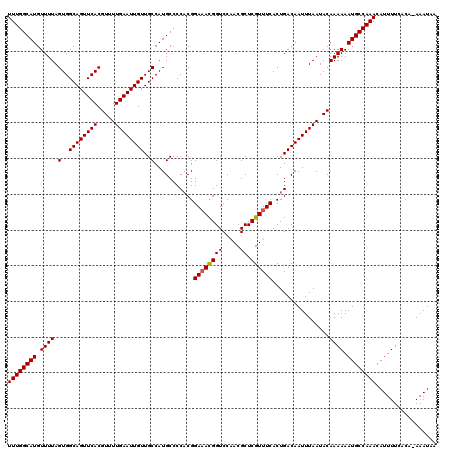
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19198093 120 + 23771897 CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCUCGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((...((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..))))))(((....)))......)))))))........ ( -39.50) >DroSec_CAF1 26446 120 + 1 CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((..(((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))))((....)).......)))))))........ ( -40.60) >DroSim_CAF1 30759 120 + 1 CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((..(((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))))((....)).......)))))))........ ( -40.60) >DroEre_CAF1 29608 120 + 1 CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((..(((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))))((....)).......)))))))........ ( -40.60) >DroYak_CAF1 54582 120 + 1 CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((..(((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))))((....)).......)))))))........ ( -40.60) >DroAna_CAF1 19659 119 + 1 UUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGA-AUGGUCCGACGCUCGUUUCACUGAC ...(((((((..(((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))))((((-....))))..)))))))........ ( -41.00) >consensus CUCAAUGGGCUCUGGGGCAAAUAUUUUAAAUGCGCAGAUUUUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGAC ...(((((((...((((((..........((((.(((....)))))))......(..((((((((.....))))))))..)..)))))).((....)).......)))))))........ (-39.17 = -39.03 + -0.14)

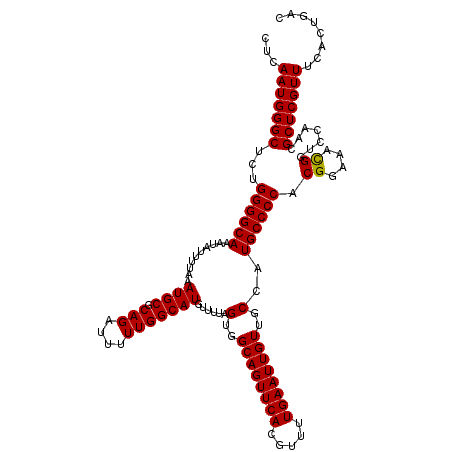
| Location | 19,198,093 – 19,198,213 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -31.09 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19198093 120 - 23771897 GUCAGUGAAACGAGCGUUGGACCGUUUCCGAGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .((((((.........(((((.....)))))(((((((((((....((((.....)))).))))).......((((((......)).))))..........))))))......)))))). ( -32.20) >DroSec_CAF1 26446 120 - 1 GUCAGUGAAACGAGCGUUGGACCGUUUCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .(((((((((((..(....)..))))))..((((((((((((....((((.....)))).))))).......((((((......)).))))..........)))))))......))))). ( -32.20) >DroSim_CAF1 30759 120 - 1 GUCAGUGAAACGAGCGUUGGACCGUUUCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .(((((((((((..(....)..))))))..((((((((((((....((((.....)))).))))).......((((((......)).))))..........)))))))......))))). ( -32.20) >DroEre_CAF1 29608 120 - 1 GUCAGUGAAACGAGCGUUGGACCGUUUCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .(((((((((((..(....)..))))))..((((((((((((....((((.....)))).))))).......((((((......)).))))..........)))))))......))))). ( -32.20) >DroYak_CAF1 54582 120 - 1 GUCAGUGAAACGAGCGUUGGACCGUUUCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .(((((((((((..(....)..))))))..((((((((((((....((((.....)))).))))).......((((((......)).))))..........)))))))......))))). ( -32.20) >DroAna_CAF1 19659 119 - 1 GUCAGUGAAACGAGCGUCGGACCAU-UCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAA .((((((..........((((....-))))((((((((((((....((((.....)))).))))).......((((((......)).))))..........))))))).....)))))). ( -32.80) >consensus GUCAGUGAAACGAGCGUUGGACCGUUUCCGUGGGGCAUGGCAACAAUUCAAAACGUGAACUGCCACUAAAACAUGCCAAAAAUCUGCGCAUUUAAAAUAUUUGCCCCAGAGCCCAUUGAG .((((((..........((((.....))))((((((((((((....((((.....)))).))))).......((((((......)).))))..........))))))).....)))))). (-31.09 = -31.12 + 0.03)

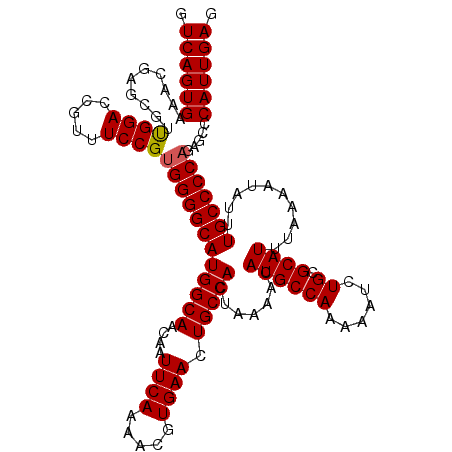

| Location | 19,198,133 – 19,198,252 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -28.57 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999890 |
| Prediction | RNA |
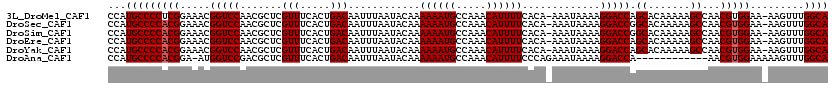
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19198133 119 + 23771897 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCUCGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAA ((((((((.((((.(..((((((((.....))))))))..)...........((((((((.....)).))))))................)))).))))))))..........-...... ( -29.20) >DroSec_CAF1 26486 119 + 1 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAA ((((((((.((((.(..((((((((.....))))))))..).......((..((((((((.....)).))))))..))............)))).))))))))..........-...... ( -29.30) >DroSim_CAF1 30799 119 + 1 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAA ((((((((.((((.(..((((((((.....))))))))..).......((..((((((((.....)).))))))..))............)))).))))))))..........-...... ( -29.30) >DroEre_CAF1 29648 119 + 1 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAA ((((((((.((((.(..((((((((.....))))))))..).......((..((((((((.....)).))))))..))............)))).))))))))..........-...... ( -29.30) >DroYak_CAF1 54622 119 + 1 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAA ((((((((.((((.(..((((((((.....))))))))..).......((..((((((((.....)).))))))..))............)))).))))))))..........-...... ( -29.30) >DroAna_CAF1 19699 119 + 1 UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGA-AUGGUCCGACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCCCAGAAAUAA ((((((((.((((.((((....))))((.(((((((((((((((.((....)).-))))).((.....)).......)))))))))).)))))).))))))))................. ( -29.30) >consensus UUUGGCAUGUUUUAGUGGCAGUUCACGUUUUGAAUUGUUGCCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA_AAAUAA ((((((((.((((.(..((((((((.....))))))))..)...........((((((((.....)).))))))................)))).))))))))................. (-28.57 = -28.60 + 0.03)
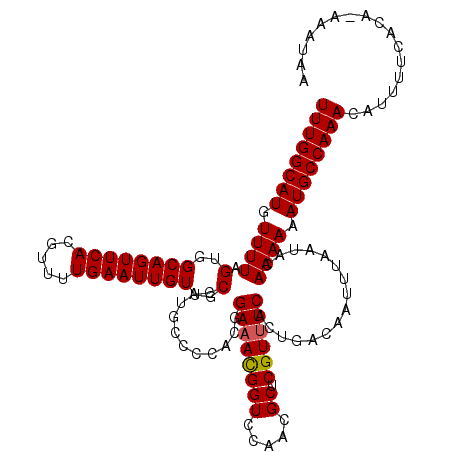
| Location | 19,198,173 – 19,198,291 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.21 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19198173 118 + 23771897 CCAUGCCCCUCGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCA ............((((((((.....)).))))))......................((((((((.(((((((.-.............................))))))-).)))))))) ( -23.31) >DroSec_CAF1 26526 118 + 1 CCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCGGCACAAAAAGCCAACGUGGAA-AAGUUUGGCA ...(((((((((.....(((((.......(((.....)))............((((((.....))))))....-........)))))(((.......)))..)))))..-......)))) ( -27.90) >DroSim_CAF1 30839 118 + 1 CCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCGGCACAAAAAGCCAACGUGGAA-AAGUUUGGCA ...(((((((((.....(((((.......(((.....)))............((((((.....))))))....-........)))))(((.......)))..)))))..-......)))) ( -27.90) >DroEre_CAF1 29688 118 + 1 CCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCA ...(((((((((.....(((((.......(((.....)))............((((((.....))))))....-........))))).((.......))...)))))..-......)))) ( -23.60) >DroYak_CAF1 54662 118 + 1 CCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCA ...(((((((((.....(((((.......(((.....)))............((((((.....))))))....-........))))).((.......))...)))))..-......)))) ( -23.60) >DroAna_CAF1 19739 107 + 1 CCAUGCCCCACGGA-AUGGUCCGACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCCCAGAAAUAAAAGGACCA------------AACGUGGAAAAAGUUUGGCA ...(((((((((..-.((((((.......(((.....)))............((((((.....)))))).............))))))------------..))))).........)))) ( -24.50) >consensus CCAUGCCCCACGGAAACGGUCCAACGCUCGUUUCACUGACAAUUUAAUACAAAAAAUGCCAAACAUUUUCACA_AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA_AAGUUUGGCA ...(((((((((.....(((((.......(((.....)))............((((((.....)))))).............))))).((.......))...))))).........)))) (-21.35 = -21.60 + 0.25)
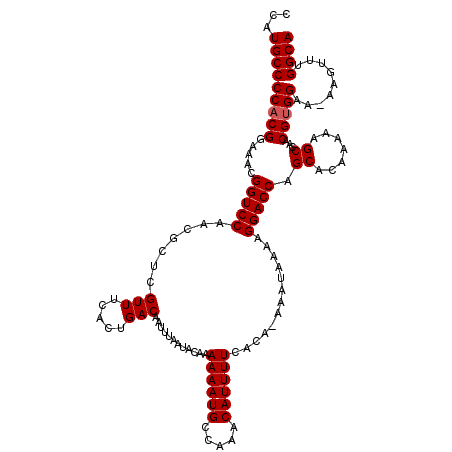
| Location | 19,198,213 – 19,198,331 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19198213 118 + 23771897 AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCAGUCGUAUAGCGUGUGUUAACCCUUUAAAAUACCCAAAAAU ...((((((((.....((((((((.(((((((.-.............................))))))-).))))))))..((.....))))))))))..................... ( -22.71) >DroSec_CAF1 26566 118 + 1 AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCGGCACAAAAAGCCAACGUGGAA-AAGUUUGGCAGUCGUAAAGCGUGUGUUAACCCUUUAAAAUACCCAAAAAU ...((((((((.....((((((((.(((((((.-........(...)(((.......)))...))))))-).))))))))..((.....))))))))))..................... ( -26.80) >DroSim_CAF1 30879 118 + 1 AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCGGCACAAAAAGCCAACGUGGAA-AAGUUUGGCAGUCGUAAAGCGUGUGUUAACCCUUUAAAAUACCCAAAAAU ...((((((((.....((((((((.(((((((.-........(...)(((.......)))...))))))-).))))))))..((.....))))))))))..................... ( -26.80) >DroEre_CAF1 29728 118 + 1 AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCAGUCGUAAAGCGUGUGUUAACCCUUUAAAAUGCCCAAAAAU ...((((((((.....((((((((.(((((((.-.............................))))))-).))))))))..((.....))))))))))..................... ( -22.71) >DroYak_CAF1 54702 118 + 1 AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA-AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA-AAGUUUGGCAGUCGUAAAGCGUGUGUUAACCCUUUAAAAUACCCAAAAAU ...((((((((.....((((((((.(((((((.-.............................))))))-).))))))))..((.....))))))))))..................... ( -22.71) >DroAna_CAF1 19778 108 + 1 AAUUUAAUACAAAAAAUGCCAAACAUUUUCCCAGAAAUAAAAGGACCA------------AACGUGGAAAAAGUUUGGCACUCGUAAACCGUGUGUUAACCCUUUAAAAUGGCCGACAAA ...((((((((.....((((((((.(((((((................------------...).)))))).))))))))..((.....)))))))))).((........))........ ( -21.11) >consensus AAUUUAAUACAAAAAAUGCCAAACAUUUUCACA_AAAUAAAAGGACCAGCACAAAAAGCCAACGUGGAA_AAGUUUGGCAGUCGUAAAGCGUGUGUUAACCCUUUAAAAUACCCAAAAAU ...((((((((.....((((((((..((((((...............................))))))...))))))))..((.....))))))))))..................... (-19.33 = -19.50 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:25 2006