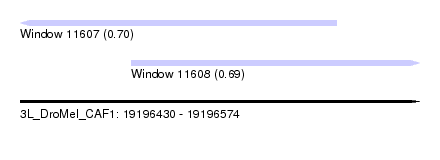
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,196,430 – 19,196,574 |
| Length | 144 |
| Max. P | 0.696141 |

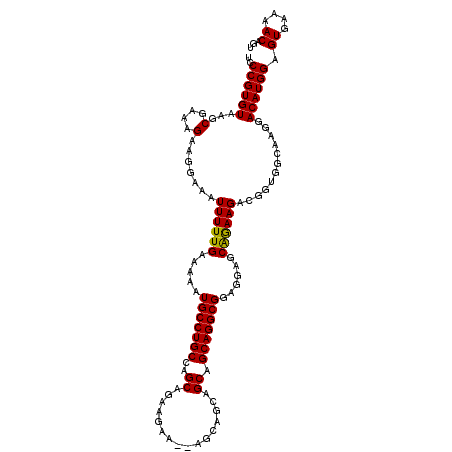
| Location | 19,196,430 – 19,196,544 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -25.16 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19196430 114 - 23771897 UCCCGUGUAAGCCAAAGAAGCAAAUUUUUGAAAAAUGCCUGCCAGCGGAAGAAGCAGCAGCAGCAGCAGGCGGAGGAGCGAAAGACGGUGGCAAGGACAUGGAGUGAAAACAGU ..((((((..((((..(..................(((((((..((.......)).((....)).)))))))......(....).)..))))....)))))).((....))... ( -33.00) >DroSec_CAF1 24698 105 - 1 UCCCGUGUAAGCGAAAGAAGGAAAUUUUUGAAAAAUGCCUGCCAGCAGAAGA---------AGCAGCAGGCGGAGGCGCAGAAGACGGUGGCAAGGACAUGGAGUGAAAACAGU ..((((((...(....).......((((((.....(((((((..((......---------.)).)))))))......))))))............)))))).((....))... ( -27.20) >DroSim_CAF1 29003 114 - 1 UUCCGUGUAAGCGAAAGAAGGAAAUUUUUGAAAAAUGCCUGCCAGCAGAAGAAUAAGCAGCAGCAGCAGGCGGAGGAGCAGAAGACGGUGGCAAGGACAUGGAGUGAAAACAGU .(((((((...(....).......((((((.....(((((((..((................)).)))))))......))))))............)))))))((....))... ( -27.59) >consensus UCCCGUGUAAGCGAAAGAAGGAAAUUUUUGAAAAAUGCCUGCCAGCAGAAGAA__AGCAGCAGCAGCAGGCGGAGGAGCAGAAGACGGUGGCAAGGACAUGGAGUGAAAACAGU ..((((((...(....).......((((((.....(((((((..((................)).)))))))......))))))............)))))).((....))... (-25.16 = -24.72 + -0.44)

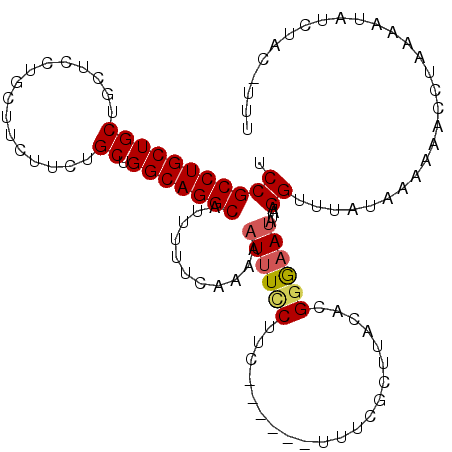
| Location | 19,196,470 – 19,196,574 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19196470 104 + 23771897 UCCGCCUGCUGCUGCUGCUGCUUCUUCCGCUGGCAGGCAUUUUUCAAAAAUUUGCUUC------UUUGGCUUACACGGGAAUAAAAGGUUUUUUAAAAAC-UGAAAUAU------UU ...(((((((((....)).((.......)).)))))))...((((.....((((.(((------(.((.....)).)))).)))).(((((.....))))-)))))...------.. ( -22.00) >DroSec_CAF1 24738 100 + 1 UCCGCCUGCUGCU---------UCUUCUGCUGGCAGGCAUUUUUCAAAAAUUUCCUUC------UUUCGCUUACACGGGAAUCUAAGGCUUAUAAAAAAAGGAAACUAUCUCG--UU ((((((((((((.---------......)).)))))))...............((((.------.(((.(......).)))...))))............)))..........--.. ( -23.60) >DroSim_CAF1 29043 110 + 1 UCCGCCUGCUGCUGCUGCUUAUUCUUCUGCUGGCAGGCAUUUUUCAAAAAUUUCCUUC------UUUCGCUUACACGGAAAUGUAAGGUUUAA-AAACAUCUAAAAUAUCUCAGUUU .(((((((((((................)).)))))))...........((((((...------............))))))....)).....-....................... ( -19.25) >DroEre_CAF1 27801 115 + 1 UCCGCCUGCUGCUGCUCCUGCUUCUUCUGCUGGCAGGCAGUUUUUAAGAAUUUUCUUCCUCAUUUUUCGCUUACACGGGAACAUACGGUUU-CAGAAG-CCUAAAAUAUCUACAUUU ...(((((((((.((....)).......)).))))))).......................(((((..((((.....(((((.....))))-)..)))-)..))))).......... ( -24.10) >DroYak_CAF1 52818 114 + 1 UCCGCCUGCUGCUGCUCCUGCUGCUUCUGCUGGCAGGCAUUUUUUAAGAAUUUCCUUCUU--UUUUUCACUUACACGGGUAUAUACGGUUUGUAUAAA-CCUAAUAUAUCUACAUUU ...(((((((((.((.......))....)).))))))).......(((((.....)))))--..............((((((((..(((((....)))-))..))))))))...... ( -27.30) >consensus UCCGCCUGCUGCUGCUCCUGCUUCUUCUGCUGGCAGGCAUUUUUCAAAAAUUUCCUUC______UUUCGCUUACACGGGAAUAUAAGGUUUAUAAAAAACCUAAAAUAUCUAC_UUU .(((((((((((................)).)))))))...........((((((.....................))))))....))............................. (-15.55 = -15.87 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:18 2006