| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,195,209 – 19,195,362 |
| Length | 153 |
| Max. P | 0.892018 |

| Location | 19,195,209 – 19,195,323 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
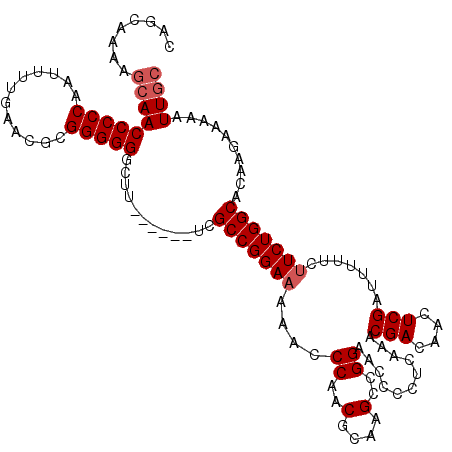
>3L_DroMel_CAF1 19195209 114 + 23771897 CAGCAAAAGCAACCCCCAAUUUUGAACGCGGGGGGCUU------UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ..((((((((..(((((............)))))))))------..(((((((....((..(....)..))...........(((....))).......)))))))..........)))) ( -29.20) >DroSec_CAF1 23485 114 + 1 CAGCAAAAGCAACCCCCAAUUUUGAACGCGGGGGGCUU------UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ..((((((((..(((((............)))))))))------..(((((((....((..(....)..))...........(((....))).......)))))))..........)))) ( -29.20) >DroSim_CAF1 27794 114 + 1 NNNNNNNNNNAACCCCCAAUUUAGAACGCGGGGGGCUU------UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ................((((((.....(((..(((.((------((....))))..)))..)))..(((((((.........(((....))).......))))))).......)))))). ( -25.69) >DroEre_CAF1 26514 114 + 1 CAGCAAAAGCAACCCCCAAUUUUGAACGCGGGGGGCUU------UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ..((((((((..(((((............)))))))))------..(((((((....((..(....)..))...........(((....))).......)))))))..........)))) ( -29.20) >DroYak_CAF1 51515 114 + 1 CGGCAAAAGCAACCCCCAAUUUUGAACGCGGGGGGCUU------UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ((((.(((((..(((((............)))))))))------).)))).......((..(....)..))...................(((((((((((.......))))))))))). ( -32.10) >DroAna_CAF1 16573 119 + 1 CAGGAAAAGCAACCCCCAAUUUUCAAUGCGGGGGGUUUUUUUUUUGGCCGGA-AAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ........(((((((((............)))))...(((((((((((((((-(...((..(....)..))...........(((....))).......))))))).))))))))))))) ( -33.50) >consensus CAGCAAAAGCAACCCCCAAUUUUGAACGCGGGGGGCUU______UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGC ........(((((((((............)))))............(((((((....((..(....)..))...........(((....))).......)))))))..........)))) (-25.32 = -25.82 + 0.50)


| Location | 19,195,247 – 19,195,362 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19195247 115 + 23771897 ----UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA-AAGAAAAACAAUUUUCUCUGCUCU ----..(((((((....((..(....)..))...........(((....))).......))))))).......................((((..-.((((((....)))))).)))).. ( -20.40) >DroSec_CAF1 23523 115 + 1 ----UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA-AAGAAAAACAAUUUUCUCUUCUCU ----..(((((((....((..(....)..))...........(((....))).......)))))))...((((...((((..........)))).-.((((((....)))))).)))).. ( -19.50) >DroSim_CAF1 27832 115 + 1 ----UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA-AAGAAAAAAAAUUUUCUCUGCUCU ----..(((((((....((..(....)..))...........(((....))).......))))))).......................((((..-.((((((....)))))).)))).. ( -20.40) >DroEre_CAF1 26552 115 + 1 ----UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA-AAGAAAAACAAUUUUCUCUUCUUU ----..(((((((....((..(....)..))...........(((....))).......)))))))..(((((...((((..........)))).-.((((((....)))))).))))). ( -20.40) >DroYak_CAF1 51553 115 + 1 ----UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA-AAGAAAAACAAUUUUCUCUUCUUU ----..(((((((....((..(....)..))...........(((....))).......)))))))..(((((...((((..........)))).-.((((((....)))))).))))). ( -20.40) >DroAna_CAF1 16613 119 + 1 UUUUUGGCCGGA-AAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAAAAAGAAAAACAAUUUUCUCUUUUUU .(((((((.((.-....)).......(((((((.........(((....))).......)))))))............))))))).......((((((((.(((....))).)))))))) ( -22.49) >consensus ____UCGCCGGAAAAACCCAACGCAAGCCGGAACCCCUCAAACGACAACUCGAUUUUUCUUCUGGCACAAGAAAAAUUGCCGAAAUUAAAGCAAA_AAGAAAAACAAUUUUCUCUUCUCU ......(((((((....((..(....)..))...........(((....))).......)))))))..........((((..........))))...((((((....))))))....... (-17.88 = -18.05 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:16 2006