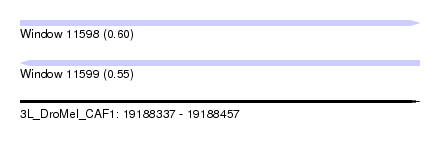
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,188,337 – 19,188,457 |
| Length | 120 |
| Max. P | 0.601008 |

| Location | 19,188,337 – 19,188,457 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.44 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19188337 120 + 23771897 CACUUGCAACGUGCAACAAAAAUUGUCAGGCUUUUAAGCCCUGAACAAAGCCAUACGCCCCCGCAGCUCGAUUAAGGGCCUCUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUUCUAU .....((.(((.((........((((((((((....)).)))).))))((((....((((....((...(((((((.....)))))))...)).....))))))))))))).))...... ( -30.70) >DroSec_CAF1 16875 108 + 1 CACUUGCAACUUGCAACAAAAAUUGUCAGGCUUUUAAGCCCUGAACAAAGCC------------AGCCCGAUUAAGGGCCUUUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUUCUAU ...((((.....))))((((..((((((((((....)).)))).))))....------------.((((......))))................))))((((((....))))))..... ( -28.80) >DroSim_CAF1 20336 108 + 1 CACUUGCAACUUGCAACAAAAAUUGUCAGGCUUUUAAGCCCUGAACAAAGCC------------AGCCCGAUUAAGGGCCUGUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUUCUAU ...((((.....))))((((..((((((((((....)).)))).))))(((.------------.((((......))))..)))...........))))((((((....))))))..... ( -30.20) >DroEre_CAF1 19698 113 + 1 CACUUU-------CAACAAAAAUUGUCAGGCUUUUGAGCCCUGAAUAAAGCCAUUCGCCCCCGAAGCCCGUUUAAGGGCCUGUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUCCUAU ......-------........((((.(((((((((((((...((((......))))((.......))..))))))))))))).))))...........(((((((....))))))).... ( -29.90) >DroYak_CAF1 44669 113 + 1 CACUUU-------CAACAAAAAUCGUCAGGCUUUUAAGCCCUGAAUAUAGCCAAACGGCCACGAAGCUCGUUUAAGGGCCUGUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUUCUAU ......-------.............(((((((((((((..........(((....)))..........)))))))))))))................(((((((....))))))).... ( -28.95) >consensus CACUUGCAAC_UGCAACAAAAAUUGUCAGGCUUUUAAGCCCUGAACAAAGCCA__CG_CC_CG_AGCCCGAUUAAGGGCCUGUUAAUCAAACUAAUUUGGGCGGCUGCUGUCGCUUCUAU ................((((.....(((((((....)).))))).....................((((......))))................))))((((((....))))))..... (-23.68 = -23.44 + -0.24)



| Location | 19,188,337 – 19,188,457 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19188337 120 - 23771897 AUAGAAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAAGAGGCCCUUAAUCGAGCUGCGGGGGCGUAUGGCUUUGUUCAGGGCUUAAAAGCCUGACAAUUUUUGUUGCACGUUGCAAGUG ......(((((.((((((((((....(((((((((((((.....)))))))))))))...))))).......(((((..(((((....)))))))))).....)))))..)))))..... ( -45.90) >DroSec_CAF1 16875 108 - 1 AUAGAAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAAAAGGCCCUUAAUCGGGCU------------GGCUUUGUUCAGGGCUUAAAAGCCUGACAAUUUUUGUUGCAAGUUGCAAGUG ......(((((.(((((.(((((((....)))).......(((((......)))))------------))).(((((..(((((....)))))))))).....)))))..)))))..... ( -34.50) >DroSim_CAF1 20336 108 - 1 AUAGAAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAACAGGCCCUUAAUCGGGCU------------GGCUUUGUUCAGGGCUUAAAAGCCUGACAAUUUUUGUUGCAAGUUGCAAGUG ......(((((.(((((.(((((((....)))).......(((((......)))))------------))).(((((..(((((....)))))))))).....)))))..)))))..... ( -34.50) >DroEre_CAF1 19698 113 - 1 AUAGGAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAACAGGCCCUUAAACGGGCUUCGGGGGCGAAUGGCUUUAUUCAGGGCUCAAAAGCCUGACAAUUUUUGUUG-------AAAGUG ......((....)).(((.(((......(((.....)))((((((......)))))).)))))).....(((((..(((((.((....)))))))((((....))))-------))))). ( -33.40) >DroYak_CAF1 44669 113 - 1 AUAGAAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAACAGGCCCUUAAACGAGCUUCGUGGCCGUUUGGCUAUAUUCAGGGCUUAAAAGCCUGACGAUUUUUGUUG-------AAAGUG .......((((((.(((((..(......(((.....))).((((.....(((.....))))))))..)))))....(((((.((....)))))))......))))))-------...... ( -27.20) >consensus AUAGAAGCGACAGCAGCCGCCCAAAUUAGUUUGAUUAACAGGCCCUUAAUCGGGCU_CG_GG_CG__UGGCUUUGUUCAGGGCUUAAAAGCCUGACAAUUUUUGUUGCA_GUUGCAAGUG ......(((((((.((((...((((....)))).......(((((......)))))............))))....(((((.((....)))))))......)))))))............ (-23.52 = -24.32 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:09 2006