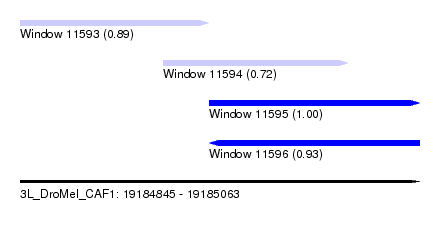
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,184,845 – 19,185,063 |
| Length | 218 |
| Max. P | 0.999758 |

| Location | 19,184,845 – 19,184,948 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19184845 103 + 23771897 GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACUGUUUUAUAUAUGUAUCUAUACUAUGG----GCAUGUAUCUGACGUAUG ........(((((.((...((((((.(((...(((......)))..(((((((.(((........))).)))))))......)))----))))))..)).))))).. ( -27.70) >DroSec_CAF1 13250 103 + 1 GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACUGUUUUAUAUAUCUAUCUAUACUGCGA----GCAUGUAUCUGGCGUAUG (((.....((((((......))))))......)))........(.((((((((.((..(((..((((........))))..))).----)).)))))))).)..... ( -24.80) >DroSim_CAF1 16779 103 + 1 GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACUGUUUUAUAUAUCUAUCUAUACUGCGA----GCAUGUAUCUGGCGUAUG (((.....((((((......))))))......)))........(.((((((((.((..(((..((((........))))..))).----)).)))))))).)..... ( -24.80) >DroEre_CAF1 16163 103 + 1 GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACAAUUUGAUAUAUCUAUCUAUACUAUAU----ACGCGUAUCUGGCGAAUA (((.....((((((......))))))......)))........(.(((((((..(((......((((....)))).........)----))..))))))).)..... ( -22.26) >DroYak_CAF1 37206 107 + 1 GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACAAUUUGAUAUAUCUAUCUAUACUAUAUAUGUACGUGUAUCUGGCAUAUA (((.....((((((......))))))......)))........(.((((((((.(((((((.((.((((......)))).)).)).))))).)))))))).)..... ( -27.70) >consensus GAGCAAUUUGCGUGGGUAAACAUGCACUAAAACUCAAUUUAGAGGCGGAUACAAGUACUGUUUUAUAUAUCUAUCUAUACUAUGA____GCAUGUAUCUGGCGUAUG (((.....((((((......))))))......)))........(.((((((((.((.........((((......))))..........)).)))))))).)..... (-20.87 = -20.83 + -0.04)


| Location | 19,184,923 – 19,185,024 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19184923 101 + 23771897 ACUAUGG----GCAUGUAUCUGACGUAUGU----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA ....(((----((......(((..(((((.----...))))).)))..(((((...(((.((((((...))))))))).))))).)))))......(((.......))) ( -25.80) >DroSec_CAF1 13328 101 + 1 ACUGCGA----GCAUGUAUCUGGCGUAUGU----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA ...((..----(((.(...((((((((...----...))))).)))..).)))(((((((((((((...))))((((.(((....)))))))...)))))))))..)). ( -30.20) >DroSim_CAF1 16857 101 + 1 ACUGCGA----GCAUGUAUCUGGCGUAUGU----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA ...((..----(((.(...((((((((...----...))))).)))..).)))(((((((((((((...))))((((.(((....)))))))...)))))))))..)). ( -30.20) >DroEre_CAF1 16241 100 + 1 ACUAUAU----ACGCGUAUCUGGCGAAUAC----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUG-GGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA .......----..(((((.((((((.....----.....))).)))....)))(((((((((((((...))))((((-(((....)))))))...)))))))))..)). ( -27.50) >DroYak_CAF1 37284 109 + 1 ACUAUAUAUGUACGUGUAUCUGGCAUAUACAUUCAUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA ...((((((....))))))((((((((..........))))).)))....((((((((((((((((...))))((((.(((....)))))))...)))))))))..))) ( -25.40) >consensus ACUAUGA____GCAUGUAUCUGGCGUAUGU____AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCA ...............(((.((((((((..........))))).)))....)))(((((((((((((...))))((((.(((....)))))))...)))))))))..... (-21.92 = -22.16 + 0.24)

| Location | 19,184,948 – 19,185,063 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19184948 115 + 23771897 U----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAGCCGCUUGAAUGAAUGAA .----.....(((.(.(((((((((((((((((((((...))))((((.(((....)))))))...)))))))))..)))))))).).)))..(..((((...))))..)......... ( -37.60) >DroSec_CAF1 13353 115 + 1 U----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAGCCACUUGAAUGAAUGAA .----.....(((.(.(((((((((((((((((((((...))))((((.(((....)))))))...)))))))))..)))))))).).)))..(..((.......))..)......... ( -32.70) >DroSim_CAF1 16882 115 + 1 U----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAGCCACUUGAAUGAAUGAA .----.....(((.(.(((((((((((((((((((((...))))((((.(((....)))))))...)))))))))..)))))))).).)))..(..((.......))..)......... ( -32.70) >DroEre_CAF1 16266 114 + 1 C----AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUG-GGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAGCCACUUGAAUGAAUGAA .----.....(((.(.(((((((((((((((((((((...))))((((-(((....)))))))...)))))))))..)))))))).).)))..(..((.......))..)......... ( -36.50) >DroYak_CAF1 37313 119 + 1 CAUUCAUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAACCACUUGAAUGAAUGAA (((((((...(((.(.(((((((((((((((((((((...))))((((.(((....)))))))...)))))))))..)))))))).).)))..(..((.......))..)))))))).. ( -38.80) >consensus U____AUCUAUGCACAGUUCUUGCAAAUUGUAUGCUGACACAGCAUUGAGGCAGGCGCCCAAUUUAAUGCGAUUUAAGCAAGAGCAGCGCAAAUUGAGCGAGCCACUUGAAUGAAUGAA ..........(((.(.(((((((((((((((((((((...))))((((.(((....)))))))...)))))))))..)))))))).).)))..(..((.......))..)......... (-32.96 = -32.96 + 0.00)

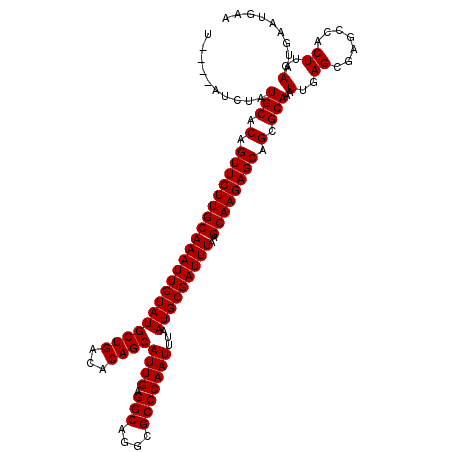
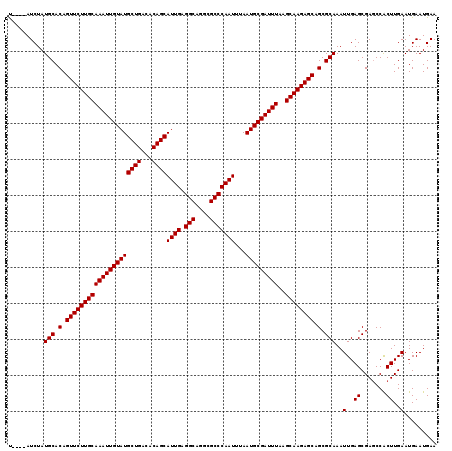
| Location | 19,184,948 – 19,185,063 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -30.29 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19184948 115 - 23771897 UUCAUUCAUUCAAGCGGCUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCCUCAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAU----A ..(((...(((..(((((.(((((((((((((..((....))......))))...)))))))))...)))...((((((...)))))).......))..)))..))).......----. ( -29.60) >DroSec_CAF1 13353 115 - 1 UUCAUUCAUUCAAGUGGCUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCCUCAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAU----A .............(.(((.(((((((((((((..((....))......))))...)))))))))...))).).((((((...))))))......((((.......)))).....----. ( -29.10) >DroSim_CAF1 16882 115 - 1 UUCAUUCAUUCAAGUGGCUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCCUCAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAU----A .............(.(((.(((((((((((((..((....))......))))...)))))))))...))).).((((((...))))))......((((.......)))).....----. ( -29.10) >DroEre_CAF1 16266 114 - 1 UUCAUUCAUUCAAGUGGCUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCC-CAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAU----G .(((((......)))))...........(((((.(.((((((................(((((....)))-))((((((...)))))).......)))))).).))))).....----. ( -30.00) >DroYak_CAF1 37313 119 - 1 UUCAUUCAUUCAAGUGGUUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCCUCAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAUGAAUG ..((((((((...(..((((((((((((((((..((....))......))))...))))))))((........((((((...)))))).......))..))))..).....)))))))) ( -33.66) >consensus UUCAUUCAUUCAAGUGGCUCGCUCAAUUUGCGCUGCUCUUGCUUAAAUCGCAUUAAAUUGGGCGCCUGCCUCAAUGCUGUGUCAGCAUACAAUUUGCAAGAACUGUGCAUAGAU____A .............(.(((.(((((((((((((..((....))......))))...)))))))))...))).).((((((...))))))......((((.......)))).......... (-28.60 = -28.44 + -0.16)

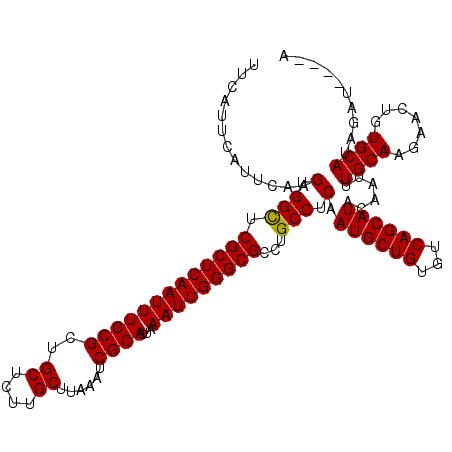
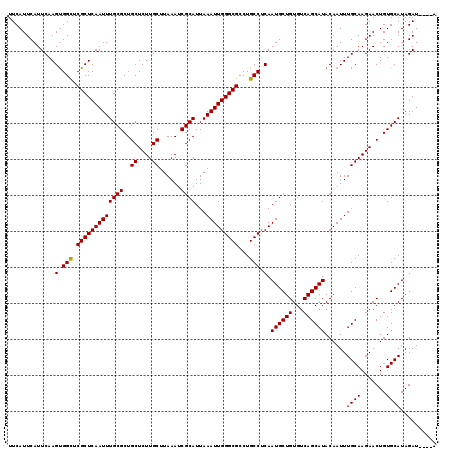
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:07 2006