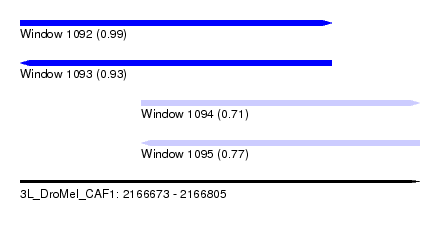
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,166,673 – 2,166,805 |
| Length | 132 |
| Max. P | 0.985815 |

| Location | 2,166,673 – 2,166,776 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2166673 103 + 23771897 AUGGGUAUUGGGAUAAUGCGUUUUCCUUGACUUGCCGCCGAAUGCUUUAAAGUGCAUCAAUUUUAAUGCAAA--U---------GCA-GCAAGCAGCAUUAACUCUUGUUGCACU ...((((..((((..........)))).....))))......(((.(((((((......))))))).)))..--(---------(((-(((((.((......))))))))))).. ( -26.70) >DroPse_CAF1 256756 100 + 1 CAGGGUAUUCG-GCAAUG-----------CCUUUCGCCCGAAUGCUUUAAAGUGCAUCAAUUUUAAUGCAAA--UAUGCAGCAAGCA-GCAAGCAGCAUUAACUUUUGCUGCGCU .((((((((((-(....(-----------(.....)))))))))))))..((((((.(((..(((((((...--..(((.....)))-.......)))))))...))).)))))) ( -30.40) >DroSec_CAF1 218671 96 + 1 AUAGGUAU-------AUGCGUUUUCCUUGACUUGCAGCCGAAUGCUUUAAAGUGCAUCAAUUUUAAUGCAAA--U---------GCA-GCAAGCAGCAUUAACUCUUGCUGCACU ...(((..-------.((((((......)))..))))))...(((.(((((((......))))))).)))..--(---------(((-(((((.((......))))))))))).. ( -27.00) >DroWil_CAF1 269875 86 + 1 CAGGGUAUUGGGC--------------------CCAUCCAAAUGUUUUAAAGUGCAUCAAUUUUAAUGCAAA--UAU-------GCAGGCAAGCAGCAUUAACUUUUGCUGCGCU ..((((.....))--------------------))...............((((((.(((..(((((((...--..(-------((......))))))))))...))).)))))) ( -23.00) >DroAna_CAF1 214375 103 + 1 CAGGGUAUC-GGAUAAUGCUUUCGCUUUGACUCGCAUCCGAUUGCU-UAAAGUGCAUCAAUUUUAAUGCAAAUAU---------GCA-GCAAGCAGCAUUAACUUUGGCUGUGCU ((.(((...-((.(((((((...((((.(.((.((((....(((((-((((((......))))))).))))..))---------)))-))))))))))))).))...))).)).. ( -27.00) >DroPer_CAF1 236406 100 + 1 CAGGGUAUUCG-GCAAUG-----------CCUUUCGCCCGAAUGCUUUAAAGUGCAUCAAUUUUAAUGCAAA--UAUUCAGCAAGCA-GCAAGCAGCAUUAACUUUUGCUGCGCU ..((((....(-((...)-----------))....))))...((((......(((((........)))))..--.....)))).(((-(((((.((......))))))))))... ( -28.72) >consensus CAGGGUAUU_G_A_AAUG___________ACUUGCACCCGAAUGCUUUAAAGUGCAUCAAUUUUAAUGCAAA__U_________GCA_GCAAGCAGCAUUAACUUUUGCUGCGCU ...(((....................................(((.(((((((......))))))).)))......................((((((........))))))))) (-16.34 = -16.03 + -0.30)

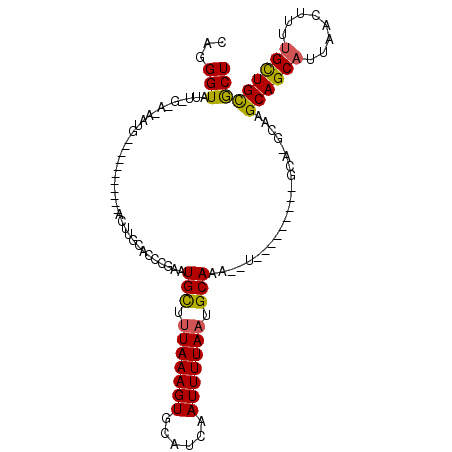

| Location | 2,166,673 – 2,166,776 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2166673 103 - 23771897 AGUGCAACAAGAGUUAAUGCUGCUUGC-UGC---------A--UUUGCAUUAAAAUUGAUGCACUUUAAAGCAUUCGGCGGCAAGUCAAGGAAAACGCAUUAUCCCAAUACCCAU ((((((((....)))..((((((((((-(..---------.--..(((((((....)))))))......))))...))))))).............))))).............. ( -25.40) >DroPse_CAF1 256756 100 - 1 AGCGCAGCAAAAGUUAAUGCUGCUUGC-UGCUUGCUGCAUA--UUUGCAUUAAAAUUGAUGCACUUUAAAGCAUUCGGGCGAAAGG-----------CAUUGC-CGAAUACCCUG ...(((((((.(((....)))..))))-))).((((.....--..(((((((....)))))))......))))...(((.....((-----------(...))-).....))).. ( -30.92) >DroSec_CAF1 218671 96 - 1 AGUGCAGCAAGAGUUAAUGCUGCUUGC-UGC---------A--UUUGCAUUAAAAUUGAUGCACUUUAAAGCAUUCGGCUGCAAGUCAAGGAAAACGCAU-------AUACCUAU ((((((((((((((....))).)))))-)))---------)--))(((((((....))))))).......((....(((.....)))..(.....)))..-------........ ( -27.60) >DroWil_CAF1 269875 86 - 1 AGCGCAGCAAAAGUUAAUGCUGCUUGCCUGC-------AUA--UUUGCAUUAAAAUUGAUGCACUUUAAAACAUUUGGAUGG--------------------GCCCAAUACCCUG ...((((((........))))))..((((..-------...--..(((((((....))))))).(((((.....))))).))--------------------))........... ( -19.20) >DroAna_CAF1 214375 103 - 1 AGCACAGCCAAAGUUAAUGCUGCUUGC-UGC---------AUAUUUGCAUUAAAAUUGAUGCACUUUA-AGCAAUCGGAUGCGAGUCAAAGCGAAAGCAUUAUCC-GAUACCCUG ((((.(((...(((....)))))))))-)((---------.((..(((((((....)))))))...))-.)).((((((((.(...)...((....))..)))))-)))...... ( -25.50) >DroPer_CAF1 236406 100 - 1 AGCGCAGCAAAAGUUAAUGCUGCUUGC-UGCUUGCUGAAUA--UUUGCAUUAAAAUUGAUGCACUUUAAAGCAUUCGGGCGAAAGG-----------CAUUGC-CGAAUACCCUG ...(((((((.(((....)))..))))-))).((((.....--..(((((((....)))))))......))))...(((.....((-----------(...))-).....))).. ( -30.92) >consensus AGCGCAGCAAAAGUUAAUGCUGCUUGC_UGC_________A__UUUGCAUUAAAAUUGAUGCACUUUAAAGCAUUCGGAUGCAAGU___________CAUU_C_C_AAUACCCUG .((((((((........))))))......................(((((((....))))))).......))........................................... (-13.25 = -13.75 + 0.50)
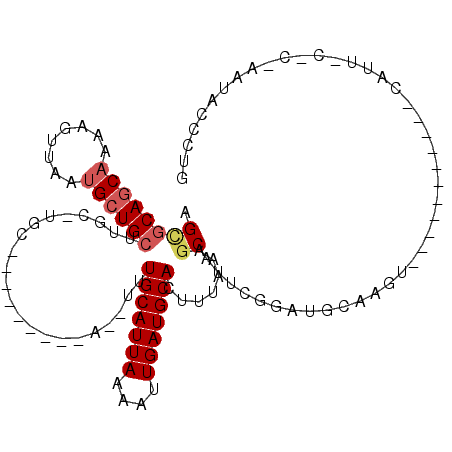
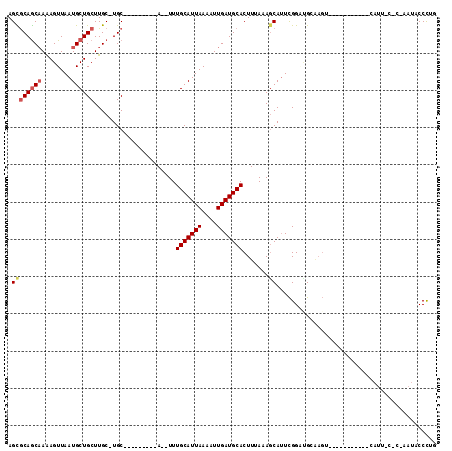
| Location | 2,166,713 – 2,166,805 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2166713 92 + 23771897 AAUGCUUUAAAGUGCAUCAAUUU--UAAUGCAAA--U---------GCA-GCAAGCAGCAUUAACUCUUGUUGCACU-----CUUAAAAAUUUACAUAAUUGCUGAAAGUC ..(((.(((((((......))))--))).)))..--.---------.((-((((((((((........))))))...-----..(((....))).....))))))...... ( -20.10) >DroVir_CAF1 240414 101 + 1 AAUGCUUUAAAGUGCAUCAAUUUAAUAAUGCAAA--CAU-------GCA-GCAAGUUGCAUUAACUUUUGCCGCGCAGGCGGCAUUAAAAUUUGCAUAAUUGCUGCGACUC .((((........)))).................--..(-------(((-((((..((((........((((((....))))))........))))...)))))))).... ( -32.19) >DroWil_CAF1 269895 95 + 1 AAUGUUUUAAAGUGCAUCAAUUU--UAAUGCAAA--UAU-------GCAGGCAAGCAGCAUUAACUUUUGCUGCGCU-----UUUACAAAUUUACAUAAUUGCUAAGAGUC ..(((...((((((((.(((..(--((((((...--..(-------((......))))))))))...))).))))))-----)).)))....................... ( -21.20) >DroMoj_CAF1 220288 101 + 1 AAUGCUUUAAAGUGCAUUAAUUUAAUAAUGCAAA--CAU-------GCA-GCAAGUUGCAUUAACUCUCGCCGCGCAGGCGGCAUUAAAAUUUGCAUAAUUGCCGCGAGUC ...((((.....(((((((......)))))))..--...-------((.-((((..((((.........(((((....))))).........))))...)))).)))))). ( -29.47) >DroAna_CAF1 214414 93 + 1 AUUGCU-UAAAGUGCAUCAAUUU--UAAUGCAAAUAU---------GCA-GCAAGCAGCAUUAACUUUGGCUGUGCU-----CUUAAAAAUUUGCAUAAUUGUUGAAAGUC ..(((.-......)))(((((..--(.((((((((..---------..(-((..(((((..........))))))))-----.......)))))))).)..)))))..... ( -20.10) >DroPer_CAF1 236434 101 + 1 AAUGCUUUAAAGUGCAUCAAUUU--UAAUGCAAA--UAUUCAGCAAGCA-GCAAGCAGCAUUAACUUUUGCUGCGCU-----CUUAAAAAUUUACAUAAUUGCUGAGAGUC ..((((......(((((......--..)))))..--.....)))).(((-(((((.((......))))))))))(((-----((((..((((.....))))..))))))). ( -27.52) >consensus AAUGCUUUAAAGUGCAUCAAUUU__UAAUGCAAA__UAU_______GCA_GCAAGCAGCAUUAACUUUUGCUGCGCU_____CUUAAAAAUUUACAUAAUUGCUGAGAGUC .((((........))))............((((.............(....)..((((((........)))))).........................))))........ (-10.88 = -10.93 + 0.06)

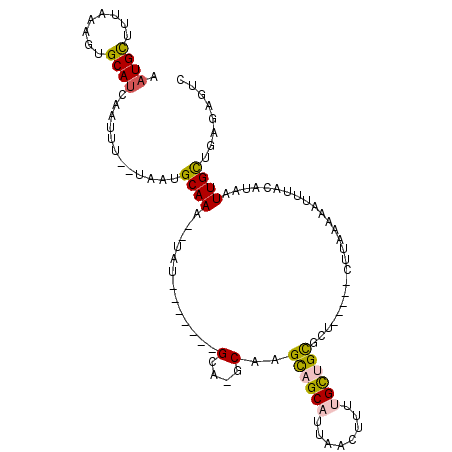
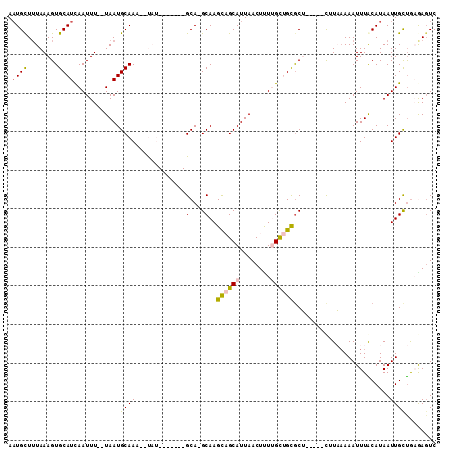
| Location | 2,166,713 – 2,166,805 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -12.24 |
| Energy contribution | -10.80 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2166713 92 - 23771897 GACUUUCAGCAAUUAUGUAAAUUUUUAAG-----AGUGCAACAAGAGUUAAUGCUGCUUGC-UGC---------A--UUUGCAUUA--AAAUUGAUGCACUUUAAAGCAUU ........((((((.....))))....((-----((((((.(((...((((((((((....-.))---------)--...))))))--)..))).))))))))...))... ( -22.90) >DroVir_CAF1 240414 101 - 1 GAGUCGCAGCAAUUAUGCAAAUUUUAAUGCCGCCUGCGCGGCAAAAGUUAAUGCAACUUGC-UGC-------AUG--UUUGCAUUAUUAAAUUGAUGCACUUUAAAGCAUU .....(((((((...((((((((((..((((((....))))))))))))..))))..))))-)))-------(((--(((((((((......)))))))......))))). ( -34.40) >DroWil_CAF1 269895 95 - 1 GACUCUUAGCAAUUAUGUAAAUUUGUAAA-----AGCGCAGCAAAAGUUAAUGCUGCUUGCCUGC-------AUA--UUUGCAUUA--AAAUUGAUGCACUUUAAAACAUU .........(((((((((((((.((((..-----.((((((((........))))))..)).)))-------).)--)))))))..--.)))))................. ( -20.80) >DroMoj_CAF1 220288 101 - 1 GACUCGCGGCAAUUAUGCAAAUUUUAAUGCCGCCUGCGCGGCGAGAGUUAAUGCAACUUGC-UGC-------AUG--UUUGCAUUAUUAAAUUAAUGCACUUUAAAGCAUU .....(((((((...((((((((((...(((((....))))).))))))..))))..))))-)))-------(((--(((((((((......)))))))......))))). ( -33.10) >DroAna_CAF1 214414 93 - 1 GACUUUCAACAAUUAUGCAAAUUUUUAAG-----AGCACAGCCAAAGUUAAUGCUGCUUGC-UGC---------AUAUUUGCAUUA--AAAUUGAUGCACUUUA-AGCAAU .....((((.....((((((((.......-----((((.(((...(((....)))))))))-)..---------..))))))))..--...))))(((......-.))).. ( -17.30) >DroPer_CAF1 236434 101 - 1 GACUCUCAGCAAUUAUGUAAAUUUUUAAG-----AGCGCAGCAAAAGUUAAUGCUGCUUGC-UGCUUGCUGAAUA--UUUGCAUUA--AAAUUGAUGCACUUUAAAGCAUU (.((((.((.((((.....)))).)).))-----)))(((((((.(((....)))..))))-))).((((.....--..(((((((--....)))))))......)))).. ( -26.12) >consensus GACUCUCAGCAAUUAUGCAAAUUUUUAAG_____AGCGCAGCAAAAGUUAAUGCUGCUUGC_UGC_______AUA__UUUGCAUUA__AAAUUGAUGCACUUUAAAGCAUU ..............((((................((((.(((....)))..))))........................(((((((......))))))).......)))). (-12.24 = -10.80 + -1.44)
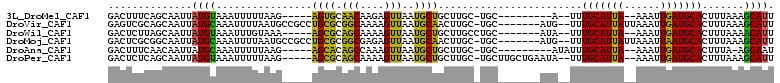
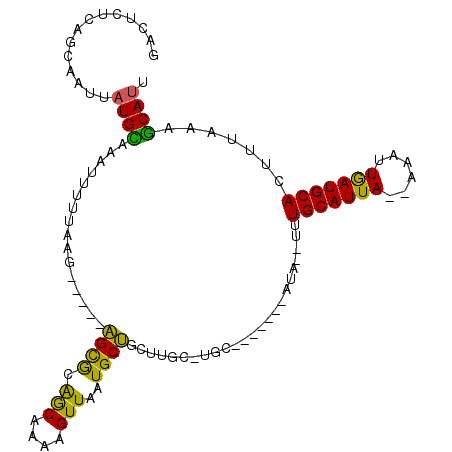
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:16 2006