| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,181,566 – 19,181,679 |
| Length | 113 |
| Max. P | 0.714902 |

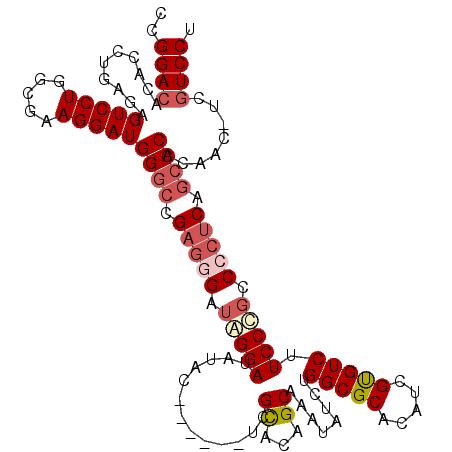
| Location | 19,181,566 – 19,181,679 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -34.28 |
| Energy contribution | -35.64 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19181566 113 + 23771897 CCGGACACACCUGAGAGUCCUGGCAAAGGAUGGGCCGAGGGAUAGGAUAUAC------UCGCACAGCAAAUAUCUGGCGCACAUCGUGUCUUCCCGCCCCUCAGCCCAAAAC-UCUUCCC ..((.....)).(((((((((.....))))(((((.(((((...(((.((((------.(((.(((.......))))))......))))..)))...))))).)))))...)-))))... ( -36.70) >DroSec_CAF1 10109 113 + 1 CCGGACACACCUGAGAGUCCUGGCGAAGGAUGGGCCGAGGGAUAGGAUAUAC------UCGCACAGCAAAUAUCUGGCGCACAUCGUGUCUUCCCGCCCCUCAGCCCACAAC-UCGUCCU ..((((..........(((((.....)))))((((.(((((...(((.((((------.(((.(((.......))))))......))))..)))...))))).)))).....-..)))). ( -39.60) >DroSim_CAF1 13228 113 + 1 CCGGACACACCUGAGAGUCCUGGCGAAGGAUGGGCCGAGGGAUGGGAUAUAC------UCGCACAGCAAAUAUCUGGCGCACAUCGUGUCUUCCCGCCCCUCAGCCCACAAC-UCGUCCU ..((((..........(((((.....)))))((((.(((((.(((((.((((------.(((.(((.......))))))......))))..))))).))))).)))).....-..)))). ( -45.20) >DroEre_CAF1 12837 119 + 1 CCGGACACACCUGAGAGUCCUGGCGAAGGAUGGGCCGC-GGAUAGGAUAUACUCGUACUCGUUCAGCAAAUAUCUGGCGCACAUCGCGUCUUCCUGCCGCUCCGCCCUCGACAUCGUCCU ..((((..........(((((.....)))))((((.((-((.(((((......((....))..............(((((.....))))).)))))))))...))))........)))). ( -39.00) >DroYak_CAF1 33734 112 + 1 CCGGACACACCUGAGAGUCCUGGCGAAGGAUGGGCCGAGGGAUAGGAUAUAC------UCGUUCAGCAAAUAUCUGGCGCACAUCGCGUCUUCCUUCCGCUCC--CCUCGACAUCGUCCU ..((((.....((((.(((((.....)))))((((.((((((..((((((..------.(.....)...))))))(((((.....))))).)))))).)))).--.)))).....)))). ( -37.60) >consensus CCGGACACACCUGAGAGUCCUGGCGAAGGAUGGGCCGAGGGAUAGGAUAUAC______UCGCACAGCAAAUAUCUGGCGCACAUCGUGUCUUCCCGCCCCUCAGCCCACAAC_UCGUCCU ..((((..........(((((.....)))))((((.(((((.(((((.............((...))........(((((.....))))).))))).))))).))))........)))). (-34.28 = -35.64 + 1.36)

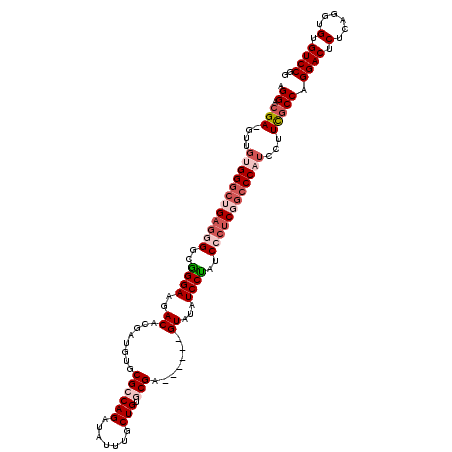
| Location | 19,181,566 – 19,181,679 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -46.52 |
| Consensus MFE | -37.48 |
| Energy contribution | -39.72 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19181566 113 - 23771897 GGGAAGA-GUUUUGGGCUGAGGGGCGGGAAGACACGAUGUGCGCCAGAUAUUUGCUGUGCGA------GUAUAUCCUAUCCCUCGGCCCAUCCUUUGCCAGGACUCUCAGGUGUGUCCGG .((.(((-(...(((((((((((..((((...(((...)))((((((.......))).))).------.....))))..)))))))))))..)))).)).((((.(......).)))).. ( -49.10) >DroSec_CAF1 10109 113 - 1 AGGACGA-GUUGUGGGCUGAGGGGCGGGAAGACACGAUGUGCGCCAGAUAUUUGCUGUGCGA------GUAUAUCCUAUCCCUCGGCCCAUCCUUCGCCAGGACUCUCAGGUGUGUCCGG .((.(((-(..((((((((((((..((((...(((...)))((((((.......))).))).------.....))))..))))))))))))..)))))).((((.(......).)))).. ( -52.90) >DroSim_CAF1 13228 113 - 1 AGGACGA-GUUGUGGGCUGAGGGGCGGGAAGACACGAUGUGCGCCAGAUAUUUGCUGUGCGA------GUAUAUCCCAUCCCUCGGCCCAUCCUUCGCCAGGACUCUCAGGUGUGUCCGG .((.(((-(..((((((((((((..((((...(((...)))((((((.......))).))).------.....))))..))))))))))))..)))))).((((.(......).)))).. ( -54.10) >DroEre_CAF1 12837 119 - 1 AGGACGAUGUCGAGGGCGGAGCGGCAGGAAGACGCGAUGUGCGCCAGAUAUUUGCUGAACGAGUACGAGUAUAUCCUAUCC-GCGGCCCAUCCUUCGCCAGGACUCUCAGGUGUGUCCGG .((((.((..((((((.((.((.((.(((....(.(((((((....(.((((((.....)))))))..))))))))..)))-)).)))).((((.....))))))))).)..)))))).. ( -38.90) >DroYak_CAF1 33734 112 - 1 AGGACGAUGUCGAGG--GGAGCGGAAGGAAGACGCGAUGUGCGCCAGAUAUUUGCUGAACGA------GUAUAUCCUAUCCCUCGGCCCAUCCUUCGCCAGGACUCUCAGGUGUGUCCGG .((((((((((((((--((.(((.........)))(((((((..(((.......))).....------)))))))...)))))))))..)))...((((.((....)).)))).)))).. ( -37.60) >consensus AGGACGA_GUUGUGGGCUGAGGGGCGGGAAGACACGAUGUGCGCCAGAUAUUUGCUGUGCGA______GUAUAUCCUAUCCCUCGGCCCAUCCUUCGCCAGGACUCUCAGGUGUGUCCGG .((.(((....((((((((((((..((((..((........((((((.......))).))).......))...))))..))))))))))))...))))).((((.(......).)))).. (-37.48 = -39.72 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:59 2006