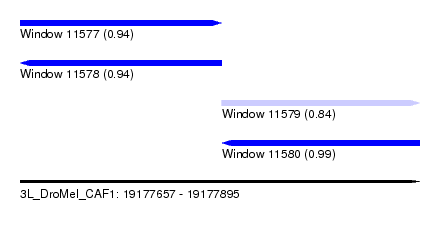
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,177,657 – 19,177,895 |
| Length | 238 |
| Max. P | 0.994957 |

| Location | 19,177,657 – 19,177,777 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19177657 120 + 23771897 GGCUAUAUUGUUGGCCGACUCCCUUGACAUUUCUAAUGGCGCAUGGCGCACAUACACCGACUGGCCAAGAGAGAUUUAUUAUCGGCUGUCAUUGUCGACGGCCAACACACACCCACACCC ((......(((((((((.(((.((((.........(((((((...)))).)))...((....)).)))).)))........(((((.......)))))))))))))).....))...... ( -37.20) >DroSec_CAF1 6124 118 + 1 GGCUAUAUUGUUGGCCGACUCCCUUGACAUUUCUAAUGGCGCAUGGCGCACAUACACCGACUGGCCAAGAGAGAUUUAUUAUCGGCUGUCAUUGUCGACGGCCAACACACACACCCAC-- ((......(((((((((.(((.((((.........(((((((...)))).)))...((....)).)))).)))........(((((.......)))))))))))))).......))..-- ( -37.02) >DroSim_CAF1 9142 118 + 1 GGCUAUAUUGUUGGCCGACUCCCUUGACAUUUCUAAUGGCGCAUGGCGCACAUACACCGACUGGCCAAGAGAGAUUUAUUAUCGGCUGUCAUUGUCGACGGCCAACACACACACCCAC-- ((......(((((((((.(((.((((.........(((((((...)))).)))...((....)).)))).)))........(((((.......)))))))))))))).......))..-- ( -37.02) >DroEre_CAF1 8893 110 + 1 GGCUAUAUUGUUGGUCGAGUCCCUUGACAUUUCUAAUGGCGCAUAGAGCAC----ACAGACUGGCCAAGAGAGAUUUAUUAUCGGCUGUCAUUGUCGAAGGCCAACACA----CCCAG-- ((......((((((((..(((....)))((((((..((((((.....)).(----(.....))))))..))))))......(((((.......))))).))))))))..----.))..-- ( -30.50) >DroYak_CAF1 11040 114 + 1 GGCUAUAUUGUUGGCCGAGUCCCUUGACAUUUCUAAUGGCGCAUAGCGCACAUACACAGACUGGCCAAAAGAGAUUUAUUAUCGGCUGUCAUUGUCGAAGGCCAACACA----CCCAC-- ((......((((((((..(((....)))((((((..((((((.....)).....((.....))))))..))))))......(((((.......))))).))))))))..----.))..-- ( -32.10) >consensus GGCUAUAUUGUUGGCCGACUCCCUUGACAUUUCUAAUGGCGCAUGGCGCACAUACACCGACUGGCCAAGAGAGAUUUAUUAUCGGCUGUCAUUGUCGACGGCCAACACACAC_CCCAC__ ........((((((((............((((((..((((((.....)).....((.....))))))..))))))......(((((.......))))).))))))))............. (-29.90 = -29.94 + 0.04)

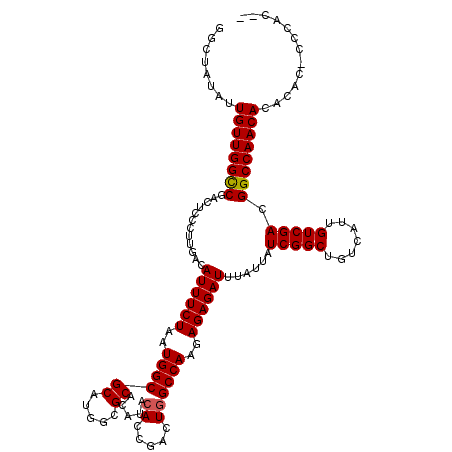

| Location | 19,177,657 – 19,177,777 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -42.54 |
| Consensus MFE | -32.40 |
| Energy contribution | -34.00 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19177657 120 - 23771897 GGGUGUGGGUGUGUGUUGGCCGUCGACAAUGACAGCCGAUAAUAAAUCUCUCUUGGCCAGUCGGUGUAUGUGCGCCAUGCGCCAUUAGAAAUGUCAAGGGAGUCGGCCAACAAUAUAGCC ......((.(((((((((((((.(..(..(((((...(((.....)))(((..((((((...(((((....))))).)).))))..)))..)))))..)..).)))))))).))))).)) ( -47.30) >DroSec_CAF1 6124 118 - 1 --GUGGGUGUGUGUGUUGGCCGUCGACAAUGACAGCCGAUAAUAAAUCUCUCUUGGCCAGUCGGUGUAUGUGCGCCAUGCGCCAUUAGAAAUGUCAAGGGAGUCGGCCAACAAUAUAGCC --..((.(((((.(((((((((.(..(..(((((...(((.....)))(((..((((((...(((((....))))).)).))))..)))..)))))..)..).)))))))))))))).)) ( -48.20) >DroSim_CAF1 9142 118 - 1 --GUGGGUGUGUGUGUUGGCCGUCGACAAUGACAGCCGAUAAUAAAUCUCUCUUGGCCAGUCGGUGUAUGUGCGCCAUGCGCCAUUAGAAAUGUCAAGGGAGUCGGCCAACAAUAUAGCC --..((.(((((.(((((((((.(..(..(((((...(((.....)))(((..((((((...(((((....))))).)).))))..)))..)))))..)..).)))))))))))))).)) ( -48.20) >DroEre_CAF1 8893 110 - 1 --CUGGG----UGUGUUGGCCUUCGACAAUGACAGCCGAUAAUAAAUCUCUCUUGGCCAGUCUGU----GUGCUCUAUGCGCCAUUAGAAAUGUCAAGGGACUCGACCAACAAUAUAGCC --..((.----(((((((....((((...........(((.....)))(((((((((...((((.----((((.....))))...))))...))))))))).))))....))))))).)) ( -32.90) >DroYak_CAF1 11040 114 - 1 --GUGGG----UGUGUUGGCCUUCGACAAUGACAGCCGAUAAUAAAUCUCUUUUGGCCAGUCUGUGUAUGUGCGCUAUGCGCCAUUAGAAAUGUCAAGGGACUCGGCCAACAAUAUAGCC --...((----(.((((((((.....(..(((((...(((.....)))(((..((((((....((((....))))..)).))))..)))..)))))..).....)))))))).....))) ( -36.10) >consensus __GUGGG_GUGUGUGUUGGCCGUCGACAAUGACAGCCGAUAAUAAAUCUCUCUUGGCCAGUCGGUGUAUGUGCGCCAUGCGCCAUUAGAAAUGUCAAGGGAGUCGGCCAACAAUAUAGCC .............(((((((((....(..(((((...(((.....)))(((..((((((...(((((....))))).)).))))..)))..)))))..)....)))))))))........ (-32.40 = -34.00 + 1.60)

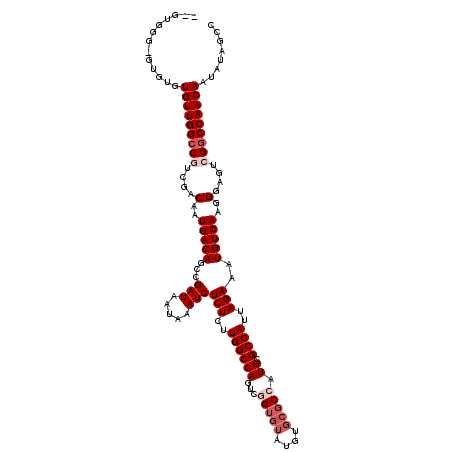

| Location | 19,177,777 – 19,177,895 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19177777 118 + 23771897 AACAUAUAUGUA--UGUAUUUAUAUGUUUUUGGCCACCAGCAAUCAUCGCCCACCCUCCUAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGCGCGAUGGCUGCUGCUUUG ((((((((....--......)))))))).........(((((..((((((....((.....)).((.(((.(((.((((.......)))).))).)).).))))))))..)))))..... ( -30.50) >DroSec_CAF1 6242 113 + 1 -----GGAUAUA--UGUAUUUAUAUGUUUUUGGCCACCAGCAAUCAUCGCCCACCCUCCCAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGCGCGAUGGCUGCUGCUUUG -----(((((((--((....)))))))))........(((((..((((((....((.....)).((.(((.(((.((((.......)))).))).)).).))))))))..)))))..... ( -30.10) >DroSim_CAF1 9260 113 + 1 -----GGAUAUA--UGUAUUUAUAUGUUUUUGGCCACCAGCAAUCAUCGCCCACCCUCCCAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGCGCGAUGGCUGGUGCUUUG -----(((((((--((....)))))))))..(((.((((((...((((((....((.....)).((.(((.(((.((((.......)))).))).)).).)))))))))))))))))... ( -32.40) >DroEre_CAF1 9003 109 + 1 -----AGAUAU------AUUUAUAUGUUUUUGGCCACCAGCAAUCAUCGCCCAUCCUCCGAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGCGCGAUGGCUGCUGCCUUG -----((((((------(....)))))))..(((....((((..((((((...(((.....)))((.(((.(((.((((.......)))).))).)).).))))))))..)))))))... ( -29.90) >DroYak_CAF1 11154 115 + 1 -----AGAUAUACUCGUAUUUAUAUGCUUUUGGCCACCAGCAAUCAUCGCCCACCCUCUCAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGUGCGAUGGCUGCUGCUUUG -----..........((((....))))..........(((((..((((((.((..(((....)))...((.(((.((((.......)))).))).))..)).))))))..)))))..... ( -25.60) >consensus _____AGAUAUA__UGUAUUUAUAUGUUUUUGGCCACCAGCAAUCAUCGCCCACCCUCCCAGGAGCCGCAAAACAUUUGUUAUAAUCAAAAGUUCUGACUGCGCGAUGGCUGCUGCUUUG .....................................(((((..((((((....((.....)).((.(((.(((.((((.......)))).))).)).).))))))))..)))))..... (-24.60 = -24.64 + 0.04)

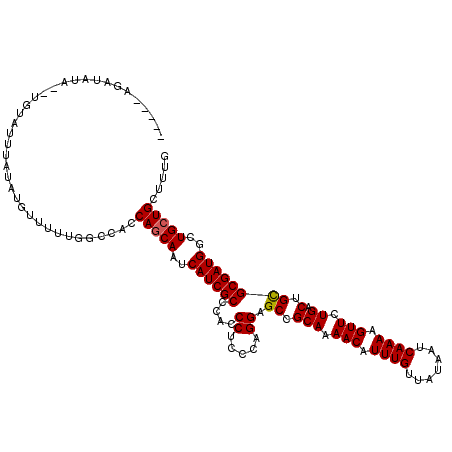

| Location | 19,177,777 – 19,177,895 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19177777 118 - 23771897 CAAAGCAGCAGCCAUCGCGCAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUAGGAGGGUGGGCGAUGAUUGCUGGUGGCCAAAAACAUAUAAAUACA--UACAUAUAUGUU .....(((((..((((((.(((.((((((.((((.......)))).)))))))..(((....)))..)).))))))..))))).........((((((((......--....)))))))) ( -35.40) >DroSec_CAF1 6242 113 - 1 CAAAGCAGCAGCCAUCGCGCAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUGGGAGGGUGGGCGAUGAUUGCUGGUGGCCAAAAACAUAUAAAUACA--UAUAUCC----- .....(((((..((((((.(((.((((((.((((.......)))).)))))))..(((....)))..)).))))))..)))))(((........))).........--.......----- ( -32.70) >DroSim_CAF1 9260 113 - 1 CAAAGCACCAGCCAUCGCGCAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUGGGAGGGUGGGCGAUGAUUGCUGGUGGCCAAAAACAUAUAAAUACA--UAUAUCC----- .....(((((((((((((.(((.((((((.((((.......)))).)))))))..(((....)))..)).))))))...)))))))....................--.......----- ( -34.50) >DroEre_CAF1 9003 109 - 1 CAAGGCAGCAGCCAUCGCGCAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUCGGAGGAUGGGCGAUGAUUGCUGGUGGCCAAAAACAUAUAAAU------AUAUCU----- ...((((.((((((((((.(((.((((((.((((.......)))).)))))))..(((....)))..)).))))))...)))).).)))..............------......----- ( -33.40) >DroYak_CAF1 11154 115 - 1 CAAAGCAGCAGCCAUCGCACAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUGAGAGGGUGGGCGAUGAUUGCUGGUGGCCAAAAGCAUAUAAAUACGAGUAUAUCU----- .....(((((..((((((...(.((((((.((((.......)))).)))))))..(.(((....))).).))))))..)))))...((.....))....................----- ( -31.30) >consensus CAAAGCAGCAGCCAUCGCGCAGUCAGAACUUUUGAUUAUAACAAAUGUUUUGCGGCUCCUGGGAGGGUGGGCGAUGAUUGCUGGUGGCCAAAAACAUAUAAAUACA__UAUAUCU_____ .....(((((..((((((.(((.((((((.((((.......)))).)))))))..(((....)))..)).))))))..)))))(((........)))....................... (-30.42 = -30.62 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:50 2006