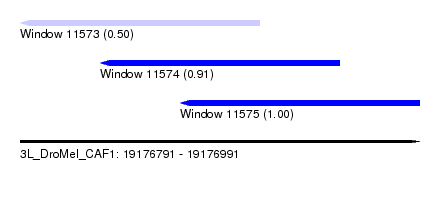
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,176,791 – 19,176,991 |
| Length | 200 |
| Max. P | 0.997966 |

| Location | 19,176,791 – 19,176,911 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19176791 120 - 23771897 GAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCGAACUUUGGACUAACAAACCGCUU ((((((((((.((((((((((((.........)))))).((....)).))))))....))))))........(((((....))))).)))).........((((......))))...... ( -33.00) >DroSec_CAF1 5308 120 - 1 GAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCGAACUUUGGACUAACAAACCGCUU ((((((((((.((((((((((((.........)))))).((....)).))))))....))))))........(((((....))))).)))).........((((......))))...... ( -33.00) >DroSim_CAF1 8345 120 - 1 GAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCAAACUUUGGACUAACAAACCGCUU ((((((((((.((((((((((((.........)))))).((....)).))))))....))))))........(((((....))))).)))).........((((......))))...... ( -33.00) >DroEre_CAF1 8048 120 - 1 GAAGGCAAGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCGAACUUUGGACUAACAAACCGCUU ..((((.....((((((((((((.........)))))).((....)).))))))...((....((((..((((((((....)))))..(((.....))).....))).))))..)))))) ( -30.90) >DroYak_CAF1 10129 120 - 1 GAACGCCAGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCGAACUUUGGACUAACAAACCGCUU ....((((...((((((((((((.........)))))).((....)).))))))..))))..(((.(((((.(((((....)))))..((((..........))))...))))).))).. ( -34.70) >consensus GAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCAGGAGUCACUUCAUCUCGAACUUUGGACUAACAAACCGCUU ((((((((((.((((((((((((.........)))))).((....)).))))))....))))))........(((((....))))).)))).........((((......))))...... (-29.60 = -30.40 + 0.80)

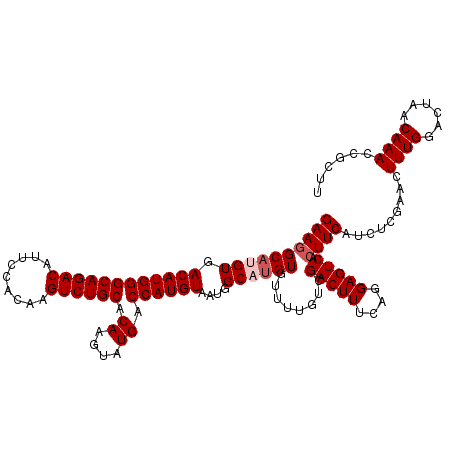
| Location | 19,176,831 – 19,176,951 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -38.44 |
| Energy contribution | -38.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19176831 120 - 23771897 UGGACUUGGACUUGGCCAUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA .(((..(.(((..((((((.((((....)))).))))))((((.((((((.((((((((((((.........)))))).((....)).))))))....)))))).))))))).)..))). ( -42.80) >DroSec_CAF1 5348 120 - 1 UGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA ..((((..((((.((((...))))))))))))(((.(.(((((.((((((.((((((((((((.........)))))).((....)).))))))....)))))).))))).).))).... ( -41.70) >DroSim_CAF1 8385 120 - 1 UGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA ..((((..((((.((((...))))))))))))(((.(.(((((.((((((.((((((((((((.........)))))).((....)).))))))....)))))).))))).).))).... ( -41.70) >DroEre_CAF1 8088 120 - 1 UGGACUUGGACGUGGCCCUUGGCUGGUUGGUCAGUGGUCGGAAGGCAAGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA ..(((..(.(((((.((....(((((((.((((.(.(((....))).).)))).((.((((((.........)))))).)).....))))).))...))))))).)...)))........ ( -39.20) >DroYak_CAF1 10169 120 - 1 UGGACUUGGACUUGGCCCUUGGCUGGUUGGUCAGUGGUUGGAACGCCAGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA ..(((..(((((..(((.((((((....)))))).)))..)...((((...((((((((((((.........)))))).((....)).))))))..))))....)))).)))........ ( -41.50) >consensus UGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCACAAGUAUGACCAUGUAAUGGCAUGUGUUUUGUCGACUUUCA ..((((..(((.(((((...))))))))))))(((.(.(((((.((((((.((((((((((((.........)))))).((....)).))))))....)))))).))))).).))).... (-38.44 = -38.88 + 0.44)

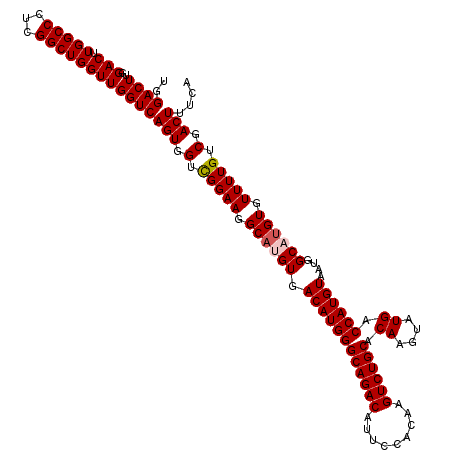
| Location | 19,176,871 – 19,176,991 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -47.90 |
| Consensus MFE | -44.42 |
| Energy contribution | -44.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
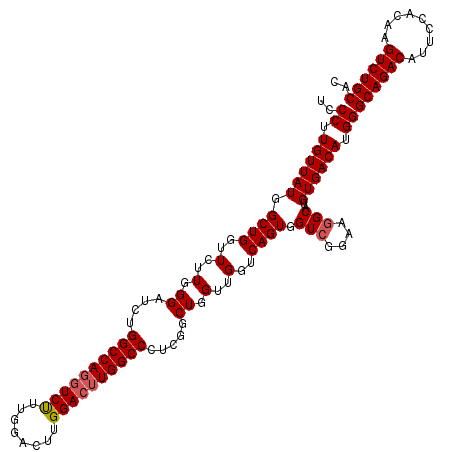
>3L_DroMel_CAF1 19176871 120 - 23771897 UCCCUUGUUAUGGCUGGUUCUUGGGAUCUGGCCAGGUCUUUGGACUUGGACUUGGCCAUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((.(((((((((..(.((....)).)..)))..((((((.((((..((((.((((...))))....))))...)))).))))))..)))))).))((((((.........)))))).. ( -49.10) >DroSec_CAF1 5388 120 - 1 UCCCUUGUUAUGGCUGGUUCUUGGGAUCUGGCCAGUUCUUUGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((.(((((((((..(.((....)).)..))).((.((((.((((..((((.((((...))))....))))...)))).))))))..)))))).))((((((.........)))))).. ( -45.50) >DroSim_CAF1 8425 120 - 1 UCCCUUGUUAUGGCUGGUUCUUGGGAUCUGGCCAGGUCUUUGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((.(((((((((..(.((....)).)..)))..((((((.((((..((((.((((...))))....))))...)))).))))))..)))))).))((((((.........)))))).. ( -49.10) >DroEre_CAF1 8128 120 - 1 UCCCUUGUUAUAGCUGGUUCUUGGGAUCUGGCCAGGUCCUUGGACUUGGACGUGGCCCUUGGCUGGUUGGUCAGUGGUCGGAAGGCAAGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((.((((((.((((..(..(.((.((.(((((.((((........)))).)))))...)))).)..)..)))).(((....)))..)))))).))((((((.........)))))).. ( -48.00) >DroYak_CAF1 10209 120 - 1 UCCCUUGUUAUAGCUGGUUCUUGGGAUCUGGCCAGGUCUUUGGACUUGGACUUGGCCCUUGGCUGGUUGGUCAGUGGUUGGAACGCCAGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((......((((..(((...(((.((.(((((((((....))))))).)).)))))..)))..))))((((.((((......)))).))))..))((((((.........)))))).. ( -47.80) >consensus UCCCUUGUUAUGGCUGGUUCUUGGGAUCUGGCCAGGUCUUUGGACUUGGACUUGGCCCUCGGCUGGUUGGUCAGUGGUCGGAAGGCAUGUGACAUGGGCAGACAUUCCACAAGUCUGCAC ..((.((((((.((((..(..(.((....((((((((((........)))))))))).....)).)..)..)))).(((....)))..)))))).))((((((.........)))))).. (-44.42 = -44.86 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:45 2006