| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,176,274 – 19,176,511 |
| Length | 237 |
| Max. P | 0.859804 |

| Location | 19,176,274 – 19,176,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19176274 120 + 23771897 UAUGGCUCUUUCUAUGCUCCGUUCUUCAUUAUUACUUAUCGCUUGGGCAAUGGCAGUCAAAUUGCUUCUGGGCCAAGCACUCUCUGUUUGGCCAUCACAUUAGAGCUGCAUAAAGUUUCC ...((..(((..(((((...(((((...............((....))(((((((((...))))).....(((((((((.....)))))))))....))))))))).))))))))...)) ( -30.00) >DroSec_CAF1 4792 120 + 1 UAUGGCUUUCUCUACGCUCCGUUCUUCAUUAUUACUUAUCGCUUGGGCAAUGGCAGUCAAAUUGCUUCUGGGCCAAACACUCUCCGUUUGGCCAUCACAUUAGAGCUGCAUAAAGUUUCC ....((...(((((.((((((((.((((...............)))).))))).))).............((((((((.......)))))))).......)))))..))........... ( -27.06) >DroSim_CAF1 7829 120 + 1 UAUGGCUUUCUCUACGCUCCGUUCUUCAUUAUUACUUAUCGCUUGGGCAAUGGCAGUCAAAUUGCUUCUGGGCCAAGCACUCUCCGUUUGACCAUCACAUUAGAGCUGCAUAAAGUUUCC ...((((((......((((.....................((((((.(...((((((...))))))....).))))))..........((.....)).....)))).....))))))... ( -23.70) >DroEre_CAF1 7538 120 + 1 UACGGCUAUUACUACGCUCCGUUCUUCAUUAUUACUUAACGCUUGGGCAAUGGCAGUCAAAUUGCUUCUCGGCCAAGCACUCUCUGUUUGGCCAUCACAUUAGAGCUGCAUAAAGUUUCC ..(((((........((((((((..............))))...))))(((((((((...))))).....(((((((((.....)))))))))....))))..)))))............ ( -29.44) >DroYak_CAF1 9618 120 + 1 UAUGGCUAUUUCUACGCUCCGUUCUUUAUUAUUACUUAACGCUUGGGCAAUGGCAGUCAAAUUGCUUCUCGGCCAAGCACUCUCUGUUUGGCCAUCACAUUAGAGCUGCAUAAAGUUUCC ...((((........((.(((..(.(((........))).)..)))))(((((((((...))))).....(((((((((.....)))))))))....))))..))))............. ( -28.50) >consensus UAUGGCUAUUUCUACGCUCCGUUCUUCAUUAUUACUUAUCGCUUGGGCAAUGGCAGUCAAAUUGCUUCUGGGCCAAGCACUCUCUGUUUGGCCAUCACAUUAGAGCUGCAUAAAGUUUCC ..(((((((......((.(((......................)))))(((((((((...))))).....((((((((.......))))))))....)))))))))))............ (-24.67 = -24.87 + 0.20)

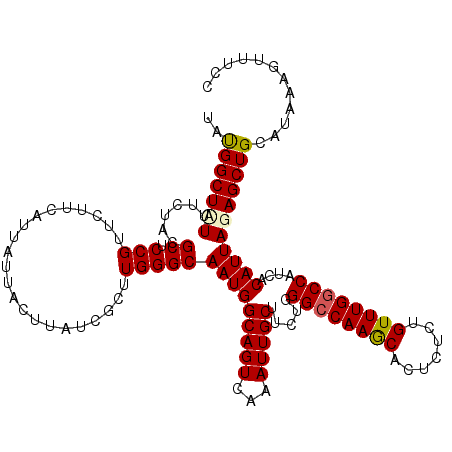

| Location | 19,176,394 – 19,176,511 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -36.79 |
| Consensus MFE | -31.87 |
| Energy contribution | -32.51 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19176394 117 - 23771897 UUUCGAUGGCGUGCUCCAA---UGUUUUCCAGCCCGAGUAACUGCGACUCGCCGGCUAACCGCACCUUUUUAACCCUUUACUUCAGCGGUUGGGGACUAAAAGCAGAACGACAUCGAACC .(((((((.((((((....---.(((..(.(((((((((.......)))))..))))((((((......................)))))))..)))....)))...))).))))))).. ( -34.85) >DroSec_CAF1 4912 117 - 1 UUUCGAUGGCGUGCUCCAA---UGUUUUCCAGCCCGAGUAACAGCGACUCGCCGGCUAACCGCACCUUUUUAACCCUUUACCUCAGCGGUUUGGGGCUAAAAGCAGAACGACAUCGAACC .(((((((.(((.((.(..---(((.....(((((((((.......)))))..))))....)))...(((((.((((..(((.....)))..)))).)))))).)).))).))))))).. ( -35.60) >DroSim_CAF1 7949 117 - 1 UUUCGAUGGCGUGCUCCAA---UGUUUUCCAGCCCGAGUAACUGCGACUCGCCUGCUAACCGCACCUUUUUAACCCUUUACCUCAGCGGUUGGGGGCUAAAAGCAGAACGACAUCGAACC .(((((((.(((.......---((((((..(((((((((.......)))).....((((((((......................))))))))))))).))))))..))).))))))).. ( -38.95) >DroEre_CAF1 7658 117 - 1 UUUGGAUGGCGUGCUCCAA---UGUUUUCCAGCCCGAGUUACGGCGACUCUCCGGCUAACCGCACCUUUUUAACCCUUUGCCUCAGCGGUUGGGGGCUGAAAGCAGAACGACAUCGAACC .((.((((.(((.......---(((((((.((((((((((.....))))).....((((((((......................))))))))))))))))))))..))).)))).)).. ( -39.85) >DroYak_CAF1 9738 120 - 1 UUUGGAUGGCGUGCUCCAACCGUGUUUUCCAGCCUGAGUUACUGCGACUCUCCGGCUAACCGCACCUUUUUAACCCUUUACCUCAGCGGUUGGGGGCUAAAAGCAGAACGACAUCGAACC .((.((((.(((.((.(....((((.....((((.(((((.....)))))...))))....))))..(((((.((((..(((.....)))..)))).)))))).)).))).)))).)).. ( -34.70) >consensus UUUCGAUGGCGUGCUCCAA___UGUUUUCCAGCCCGAGUAACUGCGACUCGCCGGCUAACCGCACCUUUUUAACCCUUUACCUCAGCGGUUGGGGGCUAAAAGCAGAACGACAUCGAACC .(((((((.(((..........((((((..(((((((((.......)))).....((((((((......................))))))))))))).))))))..))).))))))).. (-31.87 = -32.51 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:40 2006