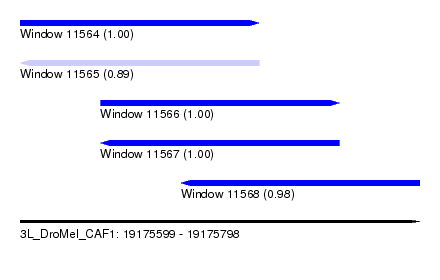
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,175,599 – 19,175,798 |
| Length | 199 |
| Max. P | 0.999737 |

| Location | 19,175,599 – 19,175,718 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
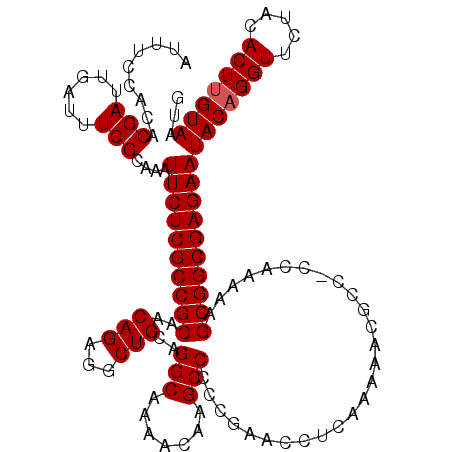
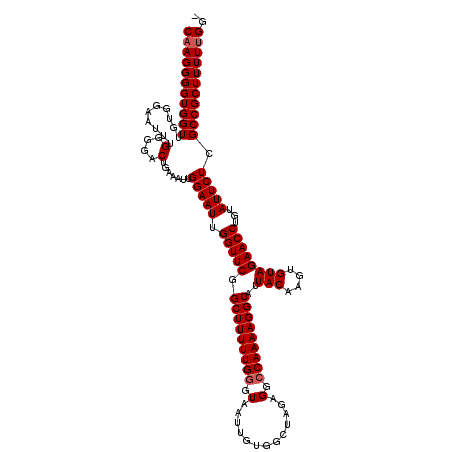
>3L_DroMel_CAF1 19175599 119 + 23771897 AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGGCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC-CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUG .........(((......)))....((((((((((....((((.((..(((.......)))..))..))))......(..-.).....))))))))))(((((((.....)))))))... ( -33.00) >DroSec_CAF1 4259 120 + 1 AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGGCUGCAGGCAAAACAAGCCCUCGAACCACAAAAACGCCCCCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUG .........(((......)))....((((((((((....((((((............)).))))(.....).................))))))))))(((((((.....)))))))... ( -31.00) >DroSim_CAF1 7162 120 + 1 AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGGCUGCAGGCAAAACAAGCCCUCGAACCACAAAAACGCCCCCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUG .........(((......)))....((((((((((....((((((............)).))))(.....).................))))))))))(((((((.....)))))))... ( -31.00) >DroEre_CAF1 6864 119 + 1 AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGUCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC-CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUG .........(((......)))....((((((((((..(((...)))..(((.......)))...................-.......))))))))))(((((((.....)))))))... ( -30.10) >DroYak_CAF1 8847 119 + 1 AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGUCUGCAGGCGAAACAAGCCCCCGAACCUCAAAAACGCC-CCGAAAAGCGGCGAGAAUACUGGUUCUACACUUGUAAUG ....(((..(((......)))...(((((((((((....(((..((..(((.......)))..))...))).....((..-.))....)))))))))))..)))...(((....)))... ( -29.80) >consensus AUUUCCACACGAUUGAUUUCGCAAAUUCUCGCCGCAACAGAGGCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC_CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUG .........(((......)))....((((((((((..(((...)))..(((.......)))...........................))))))))))(((((((.....)))))))... (-29.50 = -29.70 + 0.20)

| Location | 19,175,599 – 19,175,718 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -35.94 |
| Energy contribution | -36.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19175599 119 - 23771897 CAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGCCUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU ..((((....))))..((...(((((((((((((....)-)))((....((((((.(.((((.......)))).)))))))....)))))))))))..((((......))))..)).... ( -41.20) >DroSec_CAF1 4259 120 - 1 CAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGGGGCGUUUUUGUGGUUCGAGGGCUUGUUUUGCCUGCAGCCUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU ..((((....))))..((...(((((((((((....(((((((....(((.....)))((((.......))))...)))))))..)))))))))))..((((......))))..)).... ( -40.20) >DroSim_CAF1 7162 120 - 1 CAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGGGGCGUUUUUGUGGUUCGAGGGCUUGUUUUGCCUGCAGCCUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU ..((((....))))..((...(((((((((((....(((((((....(((.....)))((((.......))))...)))))))..)))))))))))..((((......))))..)).... ( -40.20) >DroEre_CAF1 6864 119 - 1 CAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGACUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU .....(((((.(.((((((.((....((((.(.....).-))))....))))))))..).)))))(((((((.((((......)))))))))))....((((......))))........ ( -36.90) >DroYak_CAF1 8847 119 - 1 CAUUACAAGUGUAGAACCAGUAUUCUCGCCGCUUUUCGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUCGCCUGCAGACUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU ..((((....))))..(((..(((((((((((....(((-(((......(.....)(.((((.......)))).).).)))))..)))))))))))..((((......)))).))).... ( -37.90) >consensus CAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG_GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGCCUCUGUUGCGGCGAGAAUUUGCGAAAUCAAUCGUGUGGAAAU .....(((((.(.(((((........((((.((....)).)))).......)))))..).)))))(((((((.(((((....))))))))))))....((((......))))........ (-35.94 = -36.18 + 0.24)

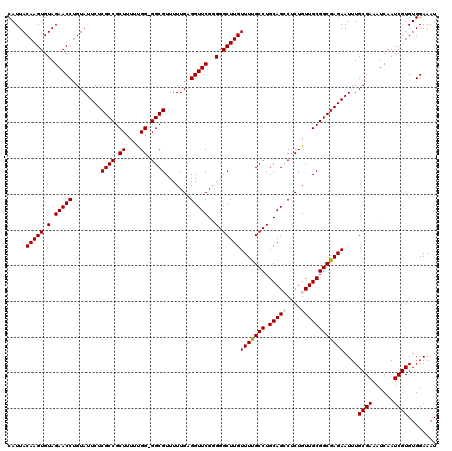
| Location | 19,175,639 – 19,175,758 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.32 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
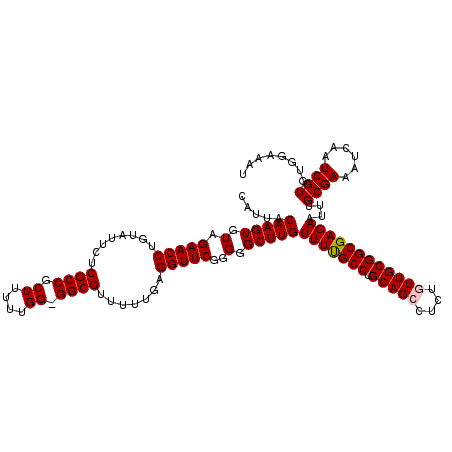
>3L_DroMel_CAF1 19175639 119 + 23771897 AGGCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC-CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUGCCUUUUGGCCUCUAGCCAUAAUUACCCAAAAAGCCGAACC .((((..(((((..(((((.....((((((......((((-.(.....).)))).......))))))....)))))..)))))..((((.....)))).............))))..... ( -34.92) >DroSec_CAF1 4299 120 + 1 AGGCUGCAGGCAAAACAAGCCCUCGAACCACAAAAACGCCCCCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUGCCUUUUGGCCUCUAGCCACAAUUACCCAAAAAGCCGAACC .((((..(((((..(((((.....(((((.......((((.(......).))))........)))))....)))))..)))))..((((.....)))).............))))..... ( -32.96) >DroSim_CAF1 7202 120 + 1 AGGCUGCAGGCAAAACAAGCCCUCGAACCACAAAAACGCCCCCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUGCCUUUUGGCCUCUAGCCACAAUUACCCAAAAAGCCGAACC .((((..(((((..(((((.....(((((.......((((.(......).))))........)))))....)))))..)))))..((((.....)))).............))))..... ( -32.96) >DroEre_CAF1 6904 119 + 1 AGUCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC-CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUGCCUUUUGGCCUCUAGCCACAAUUACCCAAAAAGCCGAACC ..((.(((((((..(((((.....((((((......((((-.(.....).)))).......))))))....)))))..)))))..((((.....))))..............)).))... ( -31.42) >DroYak_CAF1 8887 119 + 1 AGUCUGCAGGCGAAACAAGCCCCCGAACCUCAAAAACGCC-CCGAAAAGCGGCGAGAAUACUGGUUCUACACUUGUAAUGCCUUUUGCCCUCUAGCCACAAUUACCCAAAAAGCCGAACC ..((.(((((((..(((((.....(((((.......((((-.(.....).))))........)))))....)))))..)))))...((......))................)).))... ( -25.96) >consensus AGGCUGCAGGCAAAACAAGCCCCCGAACCUCAAAAACGCC_CCAAAAAGCGGCGAGAAUACAGGUUCUACACUUGUAAUGCCUUUUGGCCUCUAGCCACAAUUACCCAAAAAGCCGAACC .((((..(((((..(((((.....(((((.......((((.(......).))))........)))))....)))))..)))))..((((.....)))).............))))..... (-28.16 = -28.60 + 0.44)

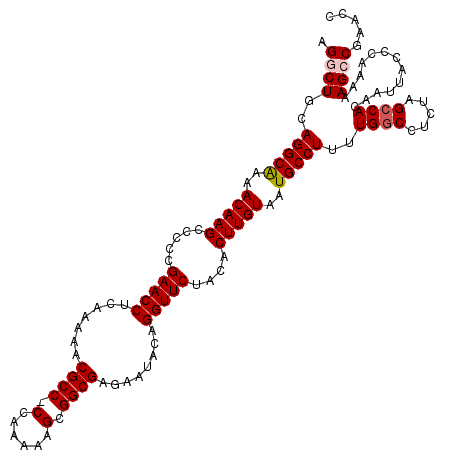
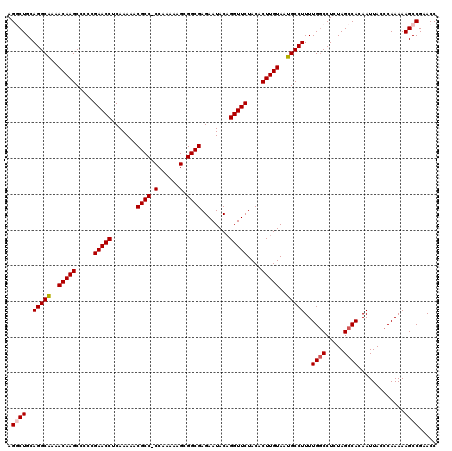
| Location | 19,175,639 – 19,175,758 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.32 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -39.18 |
| Energy contribution | -39.98 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19175639 119 - 23771897 GGUUCGGCUUUUUGGGUAAUUAUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGCCU ((((((((((((.((.((....)).)))))))))...(((((..((((((.(.((((((.((....((((.(.....).-))))....))))))))..).))))))..)))))).)))). ( -47.20) >DroSec_CAF1 4299 120 - 1 GGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGGGGCGUUUUUGUGGUUCGAGGGCUUGUUUUGCCUGCAGCCU ((((((((((((.((.((....)).)))))))))...(((((..((((((.(.(((((........((((.((....)).)))).......)))))..).))))))..)))))).)))). ( -46.46) >DroSim_CAF1 7202 120 - 1 GGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGGGGCGUUUUUGUGGUUCGAGGGCUUGUUUUGCCUGCAGCCU ((((((((((((.((.((....)).)))))))))...(((((..((((((.(.(((((........((((.((....)).)))).......)))))..).))))))..)))))).)))). ( -46.46) >DroEre_CAF1 6904 119 - 1 GGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGACU ((((.(((((((.((.((....)).)))))))))...(((((..((((((.(.((((((.((....((((.(.....).-))))....))))))))..).))))))..)))))...)))) ( -44.30) >DroYak_CAF1 8887 119 - 1 GGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGGCAAAAGGCAUUACAAGUGUAGAACCAGUAUUCUCGCCGCUUUUCGG-GGCGUUUUUGAGGUUCGGGGGCUUGUUUCGCCUGCAGACU (((..((((((((((.((....)).)))))))))...((((...((((((.(.(((((........((((.(.....).-)))).......)))))..).))))))...)))).)..))) ( -40.06) >consensus GGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG_GGCGUUUUUGAGGUUCGGGGGCUUGUUUUGCCUGCAGCCU ((((((((((((.((.((....)).)))))))))...(((((..((((((.(.(((((........((((.((....)).)))).......)))))..).))))))..)))))).)))). (-39.18 = -39.98 + 0.80)


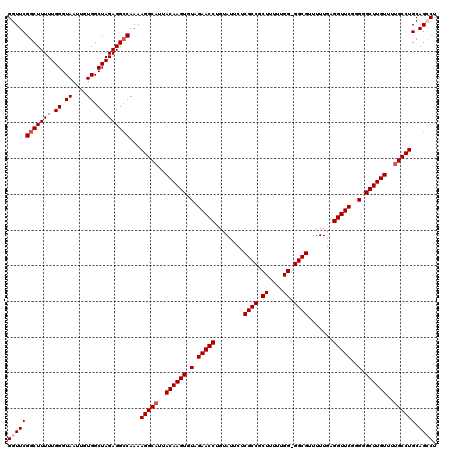
| Location | 19,175,679 – 19,175,798 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -32.88 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19175679 119 - 23771897 CAAGGGGUGGUUGUGGAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUAUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG- (((((((((((...(((((.((((..(((...((((((....))).(((((((((.(.............).)))))))))....)))...)))..)))).))))).))))))))))).- ( -36.72) >DroSec_CAF1 4339 120 - 1 CAAGGGGUGGUUGUGGAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGG (((((((((((...(((((.((((..(((...((((((....))).(((((((((.(.............).)))))))))....)))...)))..)))).))))).))))))))))).. ( -36.72) >DroSim_CAF1 7242 120 - 1 CAAGGGGUGGUUGUGGAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGGG (((((((((((...(((((.((((..(((...((((((....))).(((((((((.(.............).)))))))))....)))...)))..)))).))))).))))))))))).. ( -36.72) >DroEre_CAF1 6944 119 - 1 CAAGGGGUGGUUGUGGAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG- (((((((((((...(((((.((((..(((...((((((....))).(((((((((.(.............).)))))))))....)))...)))..)))).))))).))))))))))).- ( -36.72) >DroYak_CAF1 8927 119 - 1 CAAGGGGUGGUUGUGAAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGGCAAAAGGCAUUACAAGUGUAGAACCAGUAUUCUCGCCGCUUUUCGG- (.(((((((((.(.(((....(....)..........((((((((.(((((((((.((....)).))))))))).....((((.....)))).)))))))).))).)))))))))).).- ( -34.10) >consensus CAAGGGGUGGUUGUGGAAUUUGGGGACUGAAAUUGGAAUUGGUUCGGCUUUUUGGGUAAUUGUGGCUAGAGGCCAAAAGGCAUUACAAGUGUAGAACCUGUAUUCUCGCCGCUUUUUGG_ (((((((((((..........(....).......(((((.(((((.(((((((((.(.............).)))))))))..(((....))))))))...))))).))))))))))).. (-32.88 = -33.28 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:38 2006