
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,174,732 – 19,174,915 |
| Length | 183 |
| Max. P | 0.559629 |

| Location | 19,174,732 – 19,174,835 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
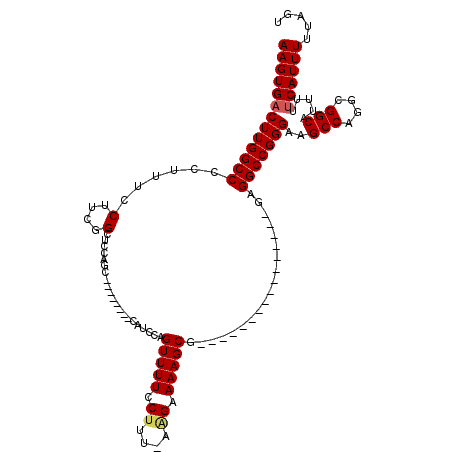
>3L_DroMel_CAF1 19174732 103 + 23771897 AAGUGACUUGGCCCCUUUCCUUCGGCUCCAGCCAUCGAGCCAUCCAGUUUUCCUUU-AAGAAAAGCG----------------GAGGCCGGGAAGCCAGGCGGCAUUUUUCAUUUUUAGU ((((((....((((((...(((((((....))).(((.(((.(((..(((((....-..)))))..)----------------))))))))))))..))).))).....))))))..... ( -34.10) >DroSec_CAF1 3379 102 + 1 AAGUGACUUGGCCCCUUUCCUUCGGCUCCAGCCAUCGAGCCAUCCAGUUUUCC--UAAAGAAAAGCG----------------GAGGCCGGGGAGCCAGGCGGCAUUUUUCAUUUUUAGU ..((..((((((((((..((((((((((........))))).....(((((.(--....).))))))----------------))))..)))).))))))..))................ ( -36.90) >DroSim_CAF1 6300 96 + 1 AAGUGACUUGGCCCCUUUCCUUCGGCUUCAGC--------CAUCCAGUUUUCCUUUAAGGAAAAGCU----------------GAGGCCGGGAAGCCAGGCGGCAUUUUUCAUUUUUAGU ((((((....((((((((((..((((((((((--------.......((((((.....)))))))))----------------)))))))))))...))).))).....))))))..... ( -36.91) >DroEre_CAF1 6029 111 + 1 AAGUGACUUGGCCCCUUUCCUUCGGCUCCAGC--------CUUCCAGUUUUCCUUU-AAGAAAAGCGAAGGCCGGGAGGCCGGGAGGCCGGGAAGCCAGUCGGCAUUUUUCAUUUUUAGU ..((((((.(((....((((..(((((((.((--------(((((.((((((((((-....)))).)))))).)))))))..))).))))))))))))))).))................ ( -44.60) >DroYak_CAF1 7934 95 + 1 AAGUGUCUUGGCCCCUUUCCUUCGGCUCGAGC--------CUUCCAGUUUUCCUUU-AAGAAAAGCG----------------GAGGCCGGGAAGCCAGUCGGCAUUUUUCAUUUUUAGU (((((((((((((..........(((....))--------).(((..(((((....-..)))))..)----------------)))))))))).(((....)))......)))))..... ( -28.00) >consensus AAGUGACUUGGCCCCUUUCCUUCGGCUCCAGC________CAUCCAGUUUUCCUUU_AAGAAAAGCG________________GAGGCCGGGAAGCCAGGCGGCAUUUUUCAUUUUUAGU (((((((((((((...........((....))..............(((((.((....)).)))))...................)))))))..(((....))).....))))))..... (-19.96 = -20.24 + 0.28)

| Location | 19,174,798 – 19,174,915 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
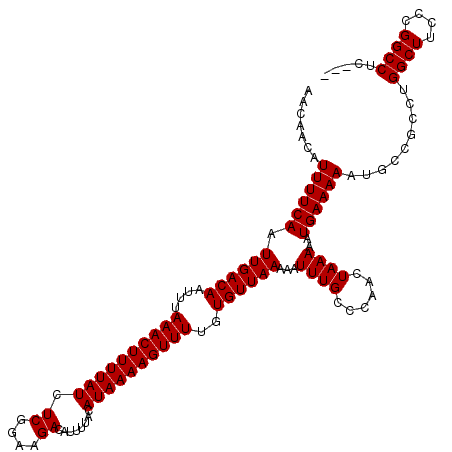
>3L_DroMel_CAF1 19174798 117 - 23771897 AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGCCCAACUAAAAAUGAAAAAUGCCGCCUGGCUUCCCGGCCUC--- .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...((((......))))..)))))).....(((.((...)).)))...--- ( -18.90) >DroSec_CAF1 3444 117 - 1 AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGCCCAACUAAAAAUGAAAAAUGCCGCCUGGCUCCCCGGCCUC--- .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...((((......))))..)))))).....(((.((...)).)))...--- ( -18.90) >DroSim_CAF1 6359 117 - 1 AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGCCCAACUAAAAAUGAAAAAUGCCGCCUGGCUUCCCGGCCUC--- .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...((((......))))..)))))).....(((.((...)).)))...--- ( -18.90) >DroEre_CAF1 6100 120 - 1 AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGCCCAACUAAAAAUGAAAAAUGCCGACUGGCUUCCCGGCCUCCCG .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...((((......))))..))))))..((((..........))))...... ( -18.80) >DroYak_CAF1 7992 116 - 1 AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGC-CAACUAAAAAUGAAAAAUGCCGACUGGCUUCCCGGCCUC--- .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...(((..-......))).))))))..((((..........))))...--- ( -18.80) >consensus AACAACAUUUUCAAUUGACAAUUUAAACUUUUAUCUCGGAAGACAUUUUACAUAAAAGUUUUGUGUUAAAAAUUUGCCCAACUAAAAAUGAAAAAUGCCGCCUGGCUUCCCGGCCUC___ .......((((((.((((((....((((((((((.((....))........))))))))))..))))))...((((......))))..)))))).........((((....))))..... (-18.40 = -18.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:34 2006