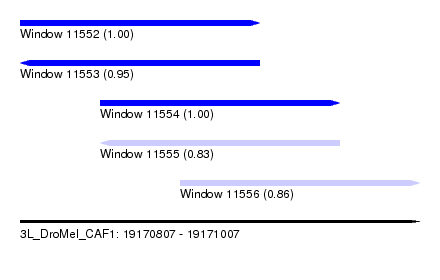
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,170,807 – 19,171,007 |
| Length | 200 |
| Max. P | 0.999357 |

| Location | 19,170,807 – 19,170,927 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170807 120 + 23771897 UGCCUGACUGCCGUGCUCCAUAUAACUAUUUGGCAAAUAAAAUAAAUUGCCCAGAGCUCUUCCGCUCCUUCUCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCA .(((.((..((((..................(((((..........)))))..((((......))))............................))))...))...))).......... ( -20.90) >DroSec_CAF1 719 118 + 1 -GCCUGACUGCCGUGUUCCAUAUAACUAUUUGGCAAAUAA-AUAAAUUGCCGAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCUGGCUUAUCUCUGGUCACACCAGCA -....(.(((..(((..(((.........(((((((....-.....)))))))((((......)))).................(((.........))).......))).)))..)))). ( -22.00) >DroSim_CAF1 3646 120 + 1 UGCCUGACUGCCGUGCUCCAUAUAACUAUUUGGCAAAUAAAAUAAAUUGCCGAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCA .(((.((..((((................(((((((..........)))))))((((......))))............................))))...))...))).......... ( -23.30) >DroEre_CAF1 3274 120 + 1 UGCCUGACUGCCGUGCUCCAUAUAACUAUUUGGCAAAUAAAAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCCCACCCACAUCUCCUCCCUUUUGCCGGCUUAUCUCUGGCCACACCAGCA .(((.((..((((.((...............(((((..........)))))..((((......))))..........................))))))...))...))).......... ( -24.10) >DroYak_CAF1 5232 119 + 1 UGCCUGACUGCCGUGCUCCAUAUAACUAUUUGGCAAAUAAAAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCUCACUCACAUCUCC-CCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCA .(((.((..((((..................(((((..........)))))..((((......))))..................-.........))))...))...))).......... ( -20.90) >consensus UGCCUGACUGCCGUGCUCCAUAUAACUAUUUGGCAAAUAAAAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCA .(((.((..((((..................(((((..........)))))..((((......))))............................))))...))...))).......... (-20.82 = -20.50 + -0.32)



| Location | 19,170,807 – 19,170,927 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170807 120 - 23771897 UGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGAGAAGGAGCGGAAGAGCUCUGGGCAAUUUAUUUUAUUUGCCAAAUAGUUAUAUGGAGCACGGCAGUCAGGCA .((((.((((.((........(((.(.....).)))...((((((.((....(((((......))))).(((((..........)))))...)).))))))))..)))).))))...... ( -34.80) >DroSec_CAF1 719 118 - 1 UGCUGGUGUGACCAGAGAUAAGCCAGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUCGGCAAUUUAU-UUAUUUGCCAAAUAGUUAUAUGGAACACGGCAGUCAGGC- .((((.((((.((........(((.........)))...((((((.((....(((((......))))).(((((.....-....)))))...)).))))))))..)))).)))).....- ( -31.70) >DroSim_CAF1 3646 120 - 1 UGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUCGGCAAUUUAUUUUAUUUGCCAAAUAGUUAUAUGGAGCACGGCAGUCAGGCA .((((.((((.((........(((.(.....).)))...((((((.((....(((((......))))).(((((..........)))))...)).))))))))..)))).))))...... ( -34.50) >DroEre_CAF1 3274 120 - 1 UGCUGGUGUGGCCAGAGAUAAGCCGGCAAAAGGGAGGAGAUGUGGGUGGGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUUUUAUUUGCCAAAUAGUUAUAUGGAGCACGGCAGUCAGGCA .((((.((((.(((...(((((((.(((............))).))).....(((((......))))).(((((..........)))))......)))).)))..)))).))))...... ( -33.50) >DroYak_CAF1 5232 119 - 1 UGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGG-GGAGAUGUGAGUGAGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUUUUAUUUGCCAAAUAGUUAUAUGGAGCACGGCAGUCAGGCA .((((.((((.((.........(((......)))-....((((((.((....(((((......))))).(((((..........)))))...)).))))))))..)))).))))...... ( -31.60) >consensus UGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUUUUAUUUGCCAAAUAGUUAUAUGGAGCACGGCAGUCAGGCA .((((.((((.((.........(((......))).....((((((.((....(((((......))))).(((((..........)))))...)).))))))))..)))).))))...... (-30.50 = -30.42 + -0.08)



| Location | 19,170,847 – 19,170,967 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -36.30 |
| Energy contribution | -37.26 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170847 120 + 23771897 AAUAAAUUGCCCAGAGCUCUUCCGCUCCUUCUCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGGAAAUGGCUGAAAGGUGGGCUCAGGGC ........((((.((((......)))).......(((((...(((.((((((((((((((.......)))).(((.......))).....)))))))))).)))...)))))....)))) ( -43.50) >DroSec_CAF1 758 119 + 1 -AUAAAUUGCCGAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCUGGCUUAUCUCUGGUCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGA -............((((......))))..((((.(((((...(((.((((((((((.(.((((..((((.....))))....))))..).)))))))))).)))...)))))....)))) ( -36.70) >DroSim_CAF1 3686 120 + 1 AAUAAAUUGCCGAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGA .............((((......))))..((((.(((((...(((.((((((((((((((.......)))).(((.......))).....)))))))))).)))...)))))....)))) ( -41.20) >DroEre_CAF1 3314 120 + 1 AAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCCCACCCACAUCUCCUCCCUUUUGCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGC ........((((.((((......)))).......(((((...((..((.((((.((((((.......)))).(((.......))).....)).)))).))..))...)))))....)))) ( -36.50) >DroYak_CAF1 5272 119 + 1 AAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCUCACUCACAUCUCC-CCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGC ........((((.((((......)))).......(((((...((.-((((((((((((((.......)))).(((.......))).....))))))))))..))...)))))....)))) ( -41.50) >consensus AAUAAAUUGCCCAGAGCUUUUCCGCUCCUUCCCACUCACAUCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGC ........((((.((((......)))).......(((((...(((.((((((((((((((.......)))).(((.......))).....)))))))))).)))...)))))....)))) (-36.30 = -37.26 + 0.96)



| Location | 19,170,847 – 19,170,967 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170847 120 - 23771897 GCCCUGAGCCCACCUUUCAGCCAUUUCCCCCAUUUCACGUUGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGAGAAGGAGCGGAAGAGCUCUGGGCAAUUUAUU ((((.((((...(((((((.(((((((.(((((((..((((.((((.....)))).....)))..)..))))))).)))))).)..)))).)))..(....).)))).))))........ ( -44.80) >DroSec_CAF1 758 119 - 1 UCCCUGAGCCCACCUUUCAGCCAUUUUCCCCAUUUCACGUUGCUGGUGUGACCAGAGAUAAGCCAGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUCGGCAAUUUAU- .......(((...(((((.((((((((((((((((..((((.((((.....)))).....)))..)..))))))).)))).))).)).)))))((((......))))..))).......- ( -36.90) >DroSim_CAF1 3686 120 - 1 UCCCUGAGCCCACCUUUCAGCCAUUUUCCCCAUUUCACGUUGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUCGGCAAUUUAUU ((((...((.(((.((((..((((((((((.(((((..((..(....)..))..))))).....))))))))))..)))).))).)))))).(((((......)))))............ ( -40.20) >DroEre_CAF1 3314 120 - 1 GCCCUGAGCCCACCUUUCAGCCAUUUUCCCCAUUUCACGUUGCUGGUGUGGCCAGAGAUAAGCCGGCAAAAGGGAGGAGAUGUGGGUGGGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUU ((((....(((((((......((((((((((........((((((((.((........)).))))))))..))).))))))).)))))))..(((((......)))))))))........ ( -47.70) >DroYak_CAF1 5272 119 - 1 GCCCUGAGCCCACCUUUCAGCCAUUUUCCCCAUUUCACGUUGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGG-GGAGAUGUGAGUGAGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUU ((((.((((((.((((((.((((((((((((((((..((((.((((.....)))).....)))..)..))))))-))))).))).))..))))).).))....)))).))))........ ( -46.70) >consensus GCCCUGAGCCCACCUUUCAGCCAUUUUCCCCAUUUCACGUUGCUGGUGUGGCCAGAGAUAAGCCGGGAAAAUGGGCGAGAUGUGAGUGGGAAGGAGCGGAAAAGCUCUGGGCAAUUUAUU .......((((((.((((..((((((((((.(((((..((..(....)..))..))))).....))))))))))..)))).)))........(((((......))))).)))........ (-35.16 = -35.80 + 0.64)

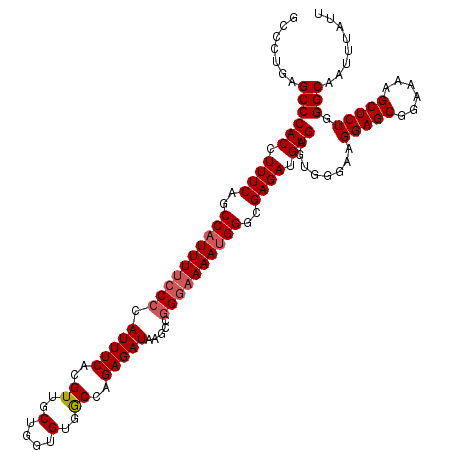

| Location | 19,170,887 – 19,171,007 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -31.84 |
| Energy contribution | -31.76 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170887 120 + 23771897 UCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGGAAAUGGCUGAAAGGUGGGCUCAGGGCUCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU ....((((((((((((((((.......)))).(((.......))).....))))))))))..(((((((((((..((((.........)))).(((......))))))).))))))))). ( -35.70) >DroSec_CAF1 797 120 + 1 UCUCGCCCAUUUUCCUGGCUUAUCUCUGGUCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGACCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU ....((((((((((((.(.((((..((((.....))))....))))..).))))))))))..(((((((((((..((((.........)))).(((......))))))).))))))))). ( -31.70) >DroSim_CAF1 3726 120 + 1 UCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGACCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU ....((((((((((((((((.......)))).(((.......))).....))))))))))..(((((((((((..((((.........)))).(((......))))))).))))))))). ( -36.20) >DroEre_CAF1 3354 120 + 1 UCUCCUCCCUUUUGCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGCCCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU ................((((.......))))...........((((((((((.(((((((((....)))((((.....)))).................)))))).))))...)))))). ( -31.90) >DroYak_CAF1 5312 119 + 1 UCUCC-CCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGCCCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU .....-((((((((((((((.......)))).(((.......))).....))))))))))..(((((((((((..((((.........)))).(((......))))))).)))))))... ( -34.80) >consensus UCUCGCCCAUUUUCCCGGCUUAUCUCUGGCCACACCAGCAACGUGAAAUGGGGAAAAUGGCUGAAAGGUGGGCUCAGGGCCCGAACAACCCUUAAAUCACAUUUUGCCUAAUUUUUCGCU ......((((((((((((((.......)))).(((.......))).....))))))))))..(((((((((((..((((.........)))).(((......))))))).)))))))... (-31.84 = -31.76 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:26 2006