
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,170,117 – 19,170,357 |
| Length | 240 |
| Max. P | 0.999945 |

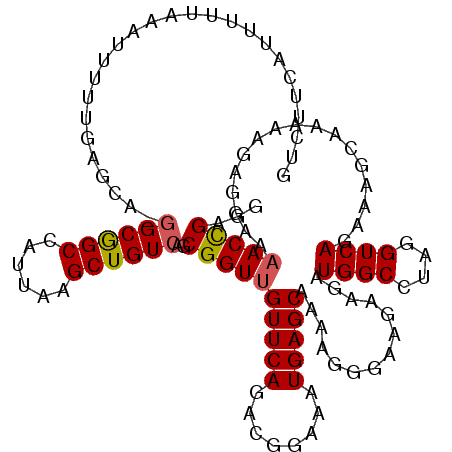
| Location | 19,170,117 – 19,170,237 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170117 120 + 23771897 GUCUUCAUUUUUAAAUUUUUGAGCAAGGCGGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGGGAAGAAGAUGGCCUAGGUCAGAAAGCAAAAAGAGAAAAACCGAGG ..((((.(((((...((((((.....((((((((((...((.((..(..(((((((........)))))))..).))))..)))))))...)))......))))))...)))))..)))) ( -29.30) >DroSec_CAF1 1 93 + 1 ---------------------------GCGGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGGGAAGAAGAUGGCCUAGGUCAGAAAGCAAAAAGAGGAAAACCGUGG ---------------------------(((((((((...((.((..(..(((((((........)))))))..).))))..)))))))....................((....)))).. ( -22.00) >DroSim_CAF1 2898 119 + 1 GUCUUCAUUUUUAAAUUUUUGAGCA-GGCGGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGGGAAGAAGAUGGCCUAGGUCAGAAAGCAAAAAGAGGAAAACCGUGG .(((((.(((((...(((((((.(.-...(((((((...((.((..(..(((((((........)))))))..).))))..)))))))..).)))))))..))))))))))......... ( -33.20) >DroEre_CAF1 2449 115 + 1 GUUUUCAUUUUUAAAUUUUUGAGCA-GGCAGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGGGAAGAAGAUGGCCUAGGUCAGAA----AAAAGAGGAAAACUGAGG ((((((.(((((...(((((((.(.-((..((((((...((.((..(..(((((((........)))))))..).))))..)))))))).).))))))----)))))).))))))..... ( -30.70) >DroYak_CAF1 4389 114 + 1 GUCUUCAUUUUUAAAUUUUUGAG-A-GGCGGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGAGAAGAAGAUGGCCUAGGUCAGAA----GAAAGAGGAAUACCGAGG .(((((..((((((....)))))-)-((((((((((...((.....(..(((((((........)))))))..)...))..)))))))...))).)))----))....((....)).... ( -28.50) >consensus GUCUUCAUUUUUAAAUUUUUGAGCA_GGCGGCCAUUAAGCUGUCAGCGGUUGUUCAGACGGAAAUGAGCAAAAGGGAAGAAGAUGGCCUAGGUCAGAAAGCAAAAAGAGGAAAACCGAGG ..........................((((((......))))))..((((((((((........)))))..............((((....)))).................)))))... (-21.34 = -21.42 + 0.08)

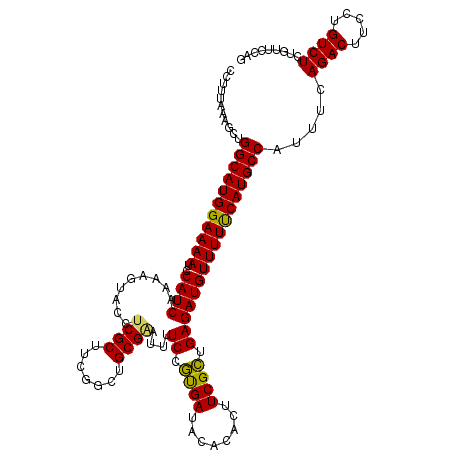
| Location | 19,170,237 – 19,170,357 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -31.30 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19170237 120 - 23771897 CCCUUUAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCCCGCUUCGUCUGCGAAUUUUCCGUGAUACACACUUCGCUGAGAUGUUUUUCAUGCCAUUUCAGACUUCCUGUCUCUGUUCCAG ..........((((((((((((..((((..........(((.......))).....((.((((........)))).))))))))))))))))))....((((.....))))......... ( -30.00) >DroSec_CAF1 94 120 - 1 CCUUUAAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCUCGCUUCGGCUGCGGAUUUUCCGUGAUACACACUUCGCUGAGAUGUUUUCCAUGCCAUUUCAGACUUCCUGUCUCUGUUCCAG ..........((((((((((((..((((...(((.....)))(((((.((((.....))))((........)))))))))))))))))))))))....((((.....))))......... ( -35.90) >DroSim_CAF1 3017 120 - 1 CCUUUAAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCUCGCUUCGGCAGCGAAUUUUCCGUGAUACACACUUCGCUGAGAUGUUUUCCAUGCCAUUUCAGACUUCCUGUCUCUGUUCCAG ..........((((((((((((..((((...........((....))((((((......(((.....)))))))))..))))))))))))))))....((((.....))))......... ( -36.50) >DroEre_CAF1 2564 120 - 1 CCUUUAAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCUCGCUUCGGCUGCGAAUUUUCCAUGAUGCACACUUCGUUGAGAUGUUUUUCAUGCCUUUUCAGACUUCCUGUCUCGGUUUCAG ......((((((((((((((((..((((.........((((.......))))....((.((((........)))).))))))))))))))))).....((((.....)))).)))))... ( -30.40) >DroYak_CAF1 4503 120 - 1 CCUUUAAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCUCGCUUCGGCUGCGAAAUUUCCACGAUGCACACUUCGUUGAGAUGUUUUUCAUGCCUUUUCAGACUUCCUGUCUCGGUUUCAG ......(((((((((((((((((..............((((.......)))).(((((.((((........)))).))))))))))))))))).....((((.....)))).)))))... ( -32.20) >consensus CCUUUAAAGCUGGCAUGGAAAAUCCAUCAAAAGUACCUCGCUUCGGCUGCGAAUUUUCCGUGAUACACACUUCGCUGAGAUGUUUUUCAUGCCAUUUCAGACUUCCUGUCUCUGUUCCAG ...........(((((((((((..((((.........((((.......))))....((.((((........)))).))))))))))))))))).....((((.....))))......... (-31.30 = -30.50 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:21 2006