
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,168,157 – 19,168,354 |
| Length | 197 |
| Max. P | 0.883940 |

| Location | 19,168,157 – 19,168,277 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -16.65 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19168157 120 + 23771897 ACACACCCAACCCAAACCCGCCACCCAGAAAAACAAACCGAGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAU .......................................(((......)))..........(((.((((.......)))).(((((((.(....))))))))...)))............ ( -15.30) >DroSim_CAF1 956 107 + 1 ACACACCCAACC-------------CAGAAAAGCAAACCGAGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAU ............-------------.......(((....(((......))).......((((((((..(............)..))))))))......(....).)))............ ( -16.80) >DroEre_CAF1 670 118 + 1 ACACACCAAAC--GACCCCGCCACCCAGGAAAACAAACCGAGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAU ...........--......((......(((.........(((......))).......((((((((..(............)..)))))))).)))..(....)..))............ ( -19.00) >DroYak_CAF1 2401 118 + 1 ACACACCAAAC--AACCCCGCCACCCAGAAAAACAAACCGAGGACAAACUCAUAAAUCUUAGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAU ...........--..........................(((......)))..........(((.((((.......)))).(((((((.(....))))))))...)))............ ( -15.50) >consensus ACACACCAAAC__AACCCCGCCACCCAGAAAAACAAACCGAGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAU .......................................(((......)))..........(((.((((.......)))).(((((((.(....))))))))...)))............ (-15.35 = -15.35 + -0.00)



| Location | 19,168,197 – 19,168,316 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.91 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -28.95 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19168197 119 + 23771897 AGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAG- ........((((...........(((((..........)))))(((((((...(((..(....)..........((((((......))))))...........)))..)))))))))))- ( -29.40) >DroSim_CAF1 983 120 + 1 AGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAGU .......(((((...........(((((..........)))))(((((((...(((..(....)..........((((((......))))))...........)))..)))))))))))) ( -30.90) >DroEre_CAF1 708 119 + 1 AGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGGAGUGUGUAAAUAACAGAAGGAGCUGCAGGAUGAG- ........((((...........(((((..........)))))(((((((...(((..(....)..........((((((......))))))...........)))..)))))))))))- ( -27.70) >DroYak_CAF1 2439 119 + 1 AGGACAAACUCAUAAAUCUUAGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGCAGUGUGUAAAUAACAGAAGGAGCUGCAGGAUGAG- ........((((...........(((((..........)))))(((((((...(((..(....)..........((((((......))))))...........)))..)))))))))))- ( -27.70) >consensus AGGACAAACUCAUAAAUCUUUGCAAGGGACUCCGACUCCCCUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAG_ ........((((...........(((((..........)))))(((((((...(((..(....)..........((((((......))))))...........)))..))))))))))). (-28.95 = -28.70 + -0.25)

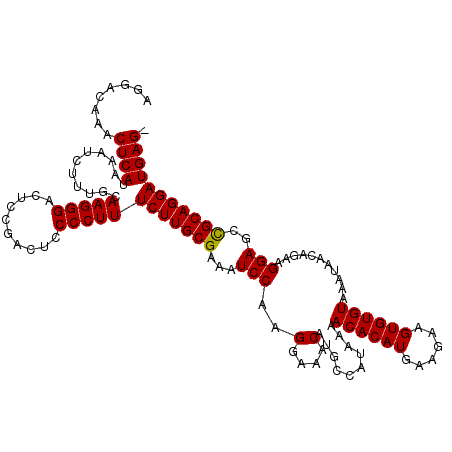
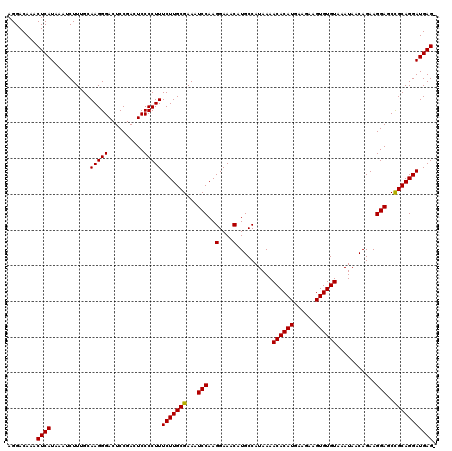
| Location | 19,168,237 – 19,168,354 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -27.52 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19168237 117 + 23771897 CUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAG---GAAGCCCCAGGAGCAUCGAGAGCACCGAAGGGGAAGUA ((((((((((...(((..(....)..........((((((......))))))...........)))..))))))).)))---....((((.((.((.......)).))...))))..... ( -34.70) >DroSim_CAF1 1023 120 + 1 CUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAGUAUGAAGCCCCAGGAGCAUCGAGAGCACCGAAGGGGAAGUA ((((((((((...(((..(....)..........((((((......))))))...........)))..))))))).))).......((((.((.((.......)).))...))))..... ( -34.00) >DroEre_CAF1 748 116 + 1 CUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGGAGUGUGUAAAUAACAGAAGGAGCUGCAGGAUGAG---GAUGCCCCAGGAGCAUCGGG-GCACCGAAGAGGAAGUA ...(((((((...(((..(....)..........((((((......))))))...........)))..)))))))...(---(.((((((.........)))-)))))............ ( -34.40) >DroYak_CAF1 2479 116 + 1 CUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGCAGUGUGUAAAUAACAGAAGGAGCUGCAGGAUGAG---GAAGCCCCAGGAGCAUCGAG-CCACCGAAGAGGAAGUA ((((((((((...(((..(....)..........((((((......))))))...........)))..))))))).)))---((.(((....).)).))..(-(..((.....))..)). ( -26.30) >consensus CUUUCUUGCGAAAUCCAAGGAAACAUGCCAUAAAACACAUGAAGAAGUGUGUAAAUAACAGAAGGAGCCGCAGGAUGAG___GAAGCCCCAGGAGCAUCGAG_GCACCGAAGAGGAAGUA ((((((((((...(((..(....)..........((((((......))))))...........)))..))))))).))).......((((.((.((.......)).))...))))..... (-27.52 = -28.03 + 0.50)

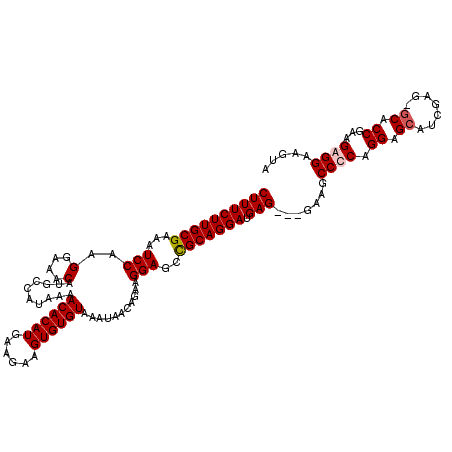

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:15 2006