
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,155,379 – 19,155,599 |
| Length | 220 |
| Max. P | 0.998067 |

| Location | 19,155,379 – 19,155,499 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -32.83 |
| Energy contribution | -32.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155379 120 + 23771897 ACAGGAUGCCUUCAUCUGCUGUUAUUGGUAUCCUGGCAUCCCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCUACCGCCACCACAGA .(((((((((.....(....).....)))))))))(((....((((((((((.(((........))).))).....))))))).)))..(((((.....................))))) ( -34.30) >DroSec_CAF1 18374 120 + 1 ACAGGAUGCCUUCAUCUGCUGUUAUUGGUAUCCUGGCAUCCCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGA .(((((((((.....(....).....)))))))))(((....((((((((((.(((........))).))).....))))))).)))................................. ( -33.00) >DroSim_CAF1 17941 120 + 1 ACAGGAUGCCUUCAUCUGCAGUUAUUGGUAUCCUGGCAUCCCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGA .(((((((((.....(....).....)))))))))(((....((((((((((.(((........))).))).....))))))).)))................................. ( -32.70) >DroEre_CAF1 18926 119 + 1 ACAGGAUGCCUUCAUCUGCUGUUAUUGGUAUCCUGGCAUCCCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCCUUCUGUUUUACCGACCCUGCCUUCC-ACCUGA ..((((((((.....(....).....))))))))((((....((((((((((.(((........))).))).....))))))).)))).........................-...... ( -32.90) >consensus ACAGGAUGCCUUCAUCUGCUGUUAUUGGUAUCCUGGCAUCCCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGA .(((((((((.....(....).....)))))))))(((....((((((((((.(((........))).))).....))))))).)))................................. (-32.83 = -32.83 + -0.00)

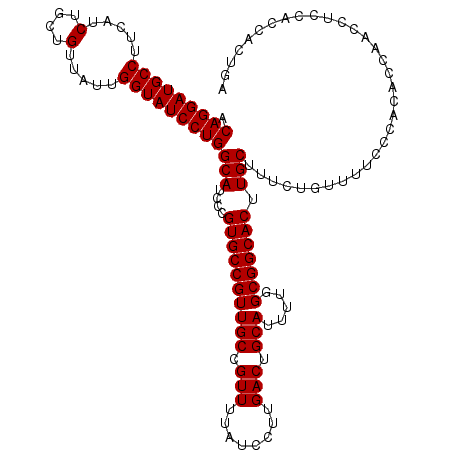
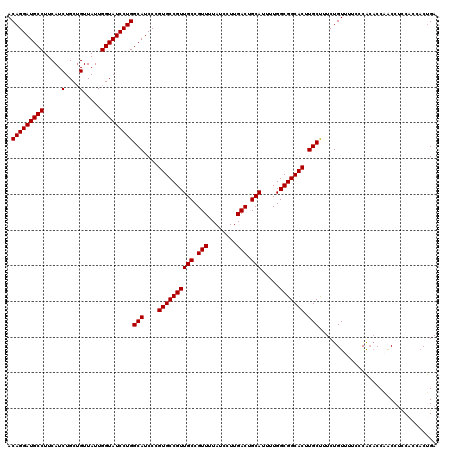
| Location | 19,155,419 – 19,155,539 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155419 120 + 23771897 CCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCUACCGCCACCACAGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((...(((.(((........))).)))....(((((....((...(((((.....................))))))).....)))))))))..(((((((((....))))))))) ( -28.30) >DroSec_CAF1 18414 120 + 1 CCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((...(((.(((........))).)))....((((((....))....((((.......................))))......))))))))..(((((((((....))))))))) ( -24.90) >DroAna_CAF1 11351 93 + 1 CUGUGCCUUUGCUGUCUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGGUUUUCCACCCU---------------------------UCGCCGCUUUGCAAAUUACUUUGCAAAA ....((...(((.(((........))).)))....((((((....))....(((....)))....---------------------------.))))))((((((((....)))))))). ( -23.60) >DroSim_CAF1 17981 120 + 1 CCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((...(((.(((........))).)))....((((((....))....((((.......................))))......))))))))..(((((((((....))))))))) ( -24.90) >DroEre_CAF1 18966 119 + 1 CCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCCUUCUGUUUUACCGACCCUGCCUUCC-ACCUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((...(((.(((........))).)))....((((((....)))...((((.....((........)).-....)))).......)))))))..(((((((((....))))))))) ( -27.60) >DroYak_CAF1 18799 119 + 1 CCGUGCCGCUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCCUUCUGUUUUACCGCCCCUGCCUUCC-CCCUGACAUUUUCCCGCAGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((((((((.(((........))).)).....)))))))).................((..((((.....-...............)))).))..(((((((((....))))))))) ( -28.95) >consensus CCGUGCCGUUGCCGUUUUAUCCUUGACUGCAUUUUGGCGGCACUUGCUUUCUGUUUUCCCACACCUACCUCCA_CACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAG ..((((((((((.(((........))).))).....))))))).......................................................(((((((((....))))))))) (-23.54 = -23.60 + 0.06)
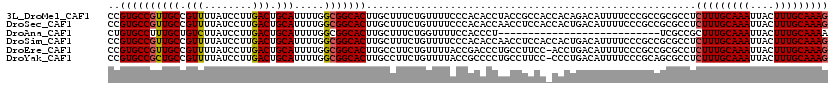

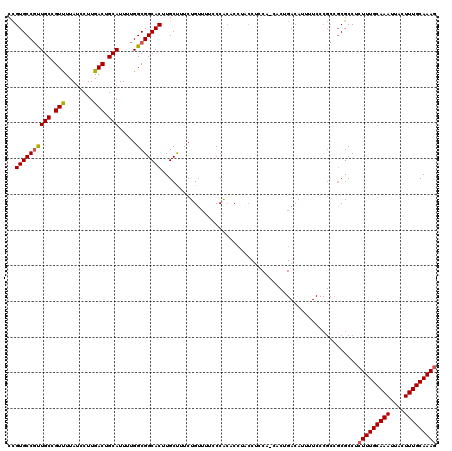
| Location | 19,155,419 – 19,155,539 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155419 120 - 23771897 CUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCUGUGGUGGCGGUAGGUGUGGGAAAACAGAAAGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAACGGCACGG (((((((((....))))))))).....((((.......((.(((((.((((((((..(((.(.....).....)))..))))))))...))))).)).........((....)))).)). ( -42.80) >DroSec_CAF1 18414 120 - 1 CUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUGGUGGAGGUUGGUGUGGGAAAACAGAAAGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAACGGCACGG (((((((((....))))))))).(.((..((.(.....((((..(((((.(..((((((((......))....((....))))))))..).)).)))..))))...).))..)).).... ( -36.10) >DroAna_CAF1 11351 93 - 1 UUUUGCAAAGUAAUUUGCAAAGCGGCGA---------------------------AGGGUGGAAAACCAGAAAGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAGACAGCAAAGGCACAG .((((((((....))))))))((((((.---------------------------..(((.....))).....((....))))))....(((.(((........))).)))...)).... ( -28.10) >DroSim_CAF1 17981 120 - 1 CUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUGGUGGAGGUUGGUGUGGGAAAACAGAAAGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAACGGCACGG (((((((((....))))))))).(.((..((.(.....((((..(((((.(..((((((((......))....((....))))))))..).)).)))..))))...).))..)).).... ( -36.10) >DroEre_CAF1 18966 119 - 1 CUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGGU-GGAAGGCAGGGUCGGUAAAACAGAAGGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAACGGCACGG (((((((((....))))))))).(.((..((.(.....((((...(-((...(((.((.(((((...(....).....)))))))....)))..)))..))))...).))..)).).... ( -37.10) >DroYak_CAF1 18799 119 - 1 CUUUGCAAAGUAAUUUGCAAAGAGGCGCUGCGGGAAAAUGUCAGGG-GGAAGGCAGGGGCGGUAAAACAGAAGGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAGCGGCACGG (((((((((....))))))))).(.((((((.(.....((((....-.((..(((..(((((((...(....).....)))))))....)))..))...))))...).)))))).).... ( -47.60) >consensus CUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUG_UGGAGGUAGGGGUGGGAAAACAGAAAGCAAGUGCCGCCAAAAUGCAGUCAAGGAUAAAACGGCAACGGCACGG (((((((((....))))))))).(((((((((......))))..................(......).........)))))(((....(((.((..........)).)))..))).... (-25.32 = -25.68 + 0.36)



| Location | 19,155,459 – 19,155,561 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155459 102 + 23771897 CACUUGCUUUCUGUUUUCCCACACCUACCGCCACCACAGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGUCACCUGCAA ....(((...(((((((..((.......(((.....................)))..((((((((((....))))))))))))..)))))))......))). ( -20.20) >DroSec_CAF1 18454 102 + 1 CACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGUCACCUGCAA ....(((...(((((((..((............................((...)).((((((((((....))))))))))))..)))))))......))). ( -18.90) >DroSim_CAF1 18021 102 + 1 CACUUGCUUUCUGUUUUCCCACACCAACCUCCACCACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGUCACCUGCAA ....(((...(((((((..((............................((...)).((((((((((....))))))))))))..)))))))......))). ( -18.90) >DroEre_CAF1 19006 101 + 1 CACUUGCCUUCUGUUUUACCGACCCUGCCUUCC-ACCUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGCCACCUGCAA ....(((...(((((((................-...............((...)).((((((((((....))))))))))....)))))))......))). ( -17.00) >DroYak_CAF1 18839 101 + 1 CACUUGCCUUCUGUUUUACCGCCCCUGCCUUCC-CCCUGACAUUUUCCCGCAGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGCCACCUGCAC ....(((...(((((((...((..((((.....-...............)))).)).((((((((((....))))))))))....)))))))......))). ( -21.15) >consensus CACUUGCUUUCUGUUUUCCCACACCUACCUCCACCACUGACAUUUUCCCGCCGCGCCUCUUUGCAAAUUACUUUGCAAAGAUGAAGAGACAGUCACCUGCAA ....(((...(((((((..((............................((...)).((((((((((....))))))))))))..)))))))......))). (-18.14 = -17.90 + -0.24)

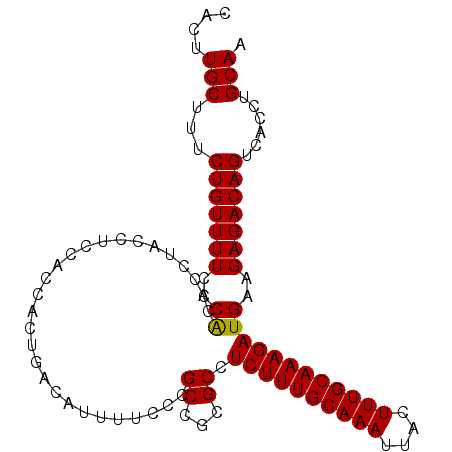
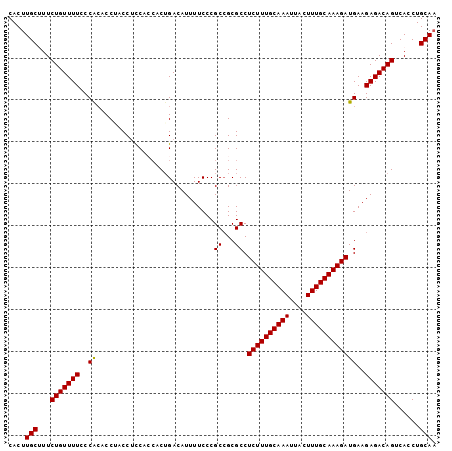
| Location | 19,155,459 – 19,155,561 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155459 102 - 23771897 UUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCUGUGGUGGCGGUAGGUGUGGGAAAACAGAAAGCAAGUG ((((((....)).((((.((.((((((((((....))))))))))(.(((.(((((......))))).))).)....))...)))).........))))... ( -34.30) >DroSec_CAF1 18454 102 - 1 UUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUGGUGGAGGUUGGUGUGGGAAAACAGAAAGCAAGUG ..((((....))))..(((((((((((((((....))))))))))..(((((((......)))).))).)))))(((..(((......)))...)))..... ( -31.10) >DroSim_CAF1 18021 102 - 1 UUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUGGUGGAGGUUGGUGUGGGAAAACAGAAAGCAAGUG ..((((....))))..(((((((((((((((....))))))))))..(((((((......)))).))).)))))(((..(((......)))...)))..... ( -31.10) >DroEre_CAF1 19006 101 - 1 UUGCAGGUGGCUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGGU-GGAAGGCAGGGUCGGUAAAACAGAAGGCAAGUG ((((..((.((((.((((((.((((((((((....))))))))))..(((((((......))))..))-)...)).)))).))))...)).....))))... ( -31.00) >DroYak_CAF1 18839 101 - 1 GUGCAGGUGGCUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCUGCGGGAAAAUGUCAGGG-GGAAGGCAGGGGCGGUAAAACAGAAGGCAAGUG .(((..((.(((((((((((.((((((((((....))))))))))..(.(((((......)).))).)-....)).)))))))))...)).....))).... ( -34.00) >consensus UUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUGUCAGUGGUGGAGGUAGGUGUGGGAAAACAGAAAGCAAGUG ..((((....)))).(((((.((((((((((....)))))))))).....((((......))))......)))))........................... (-23.02 = -23.18 + 0.16)

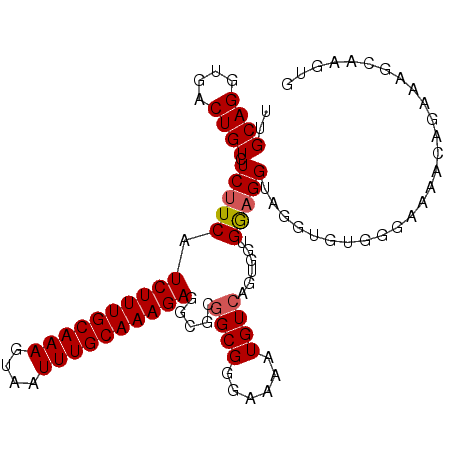
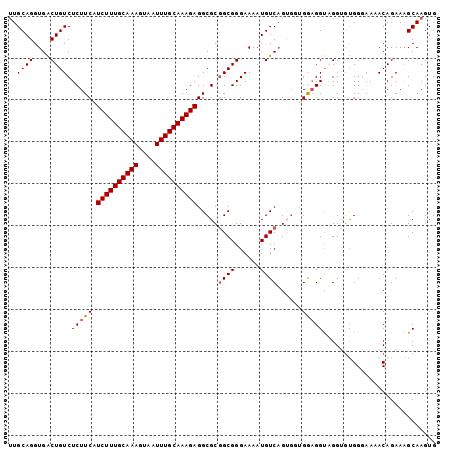
| Location | 19,155,499 – 19,155,599 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19155499 100 - 23771897 UUUACUGGGGACCACACUUUUUUUCCAAGUUUAAAAUGUUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUG ......(....).......(((((((...........(..((((....))))..)....((((((((((....))))))))))........))))))).. ( -27.30) >DroSec_CAF1 18494 100 - 1 CUUACUAGAAACCACAUCUUUUUUCCAAGUUUAAAAUGUUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUG ...................(((((((...........(..((((....))))..)....((((((((((....))))))))))........))))))).. ( -26.70) >DroSim_CAF1 18061 100 - 1 UUUACAGGAGACCGCACUUUUUUUCCAAGUUUAAAAUGUUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUG .........(.((((.((...................(..((((....))))..)....((((((((((....)))))))))))).)))))......... ( -28.60) >DroEre_CAF1 19045 98 - 1 UGCAGUGUAGACCACA--UUUUUUCCAGUUUUAAAAUGUUGCAGGUGGCUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUG ....(((.....))).--.(((((((.((...........))..((.((.(((((((......((((((....))))))))))))))).))))))))).. ( -26.40) >DroYak_CAF1 18878 96 - 1 UGCAC--UAGACCACA--UUUUUUCCAAGCUUAAAAUGGUGCAGGUGGCUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCUGCGGGAAAAUG .....--.........--.(((((((..((..........))..(..((.(((((((......((((((....)))))))))))))))..)))))))).. ( -27.90) >consensus UUUACUGGAGACCACA__UUUUUUCCAAGUUUAAAAUGUUGCAGGUGACUGUCUCUUCAUCUUUGCAAAGUAAUUUGCAAAGAGGCGCGGCGGGAAAAUG ...................(((((((...........(..((((....))))..)....((((((((((....))))))))))........))))))).. (-25.24 = -25.24 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:54 2006