
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,153,790 – 19,154,019 |
| Length | 229 |
| Max. P | 0.997782 |

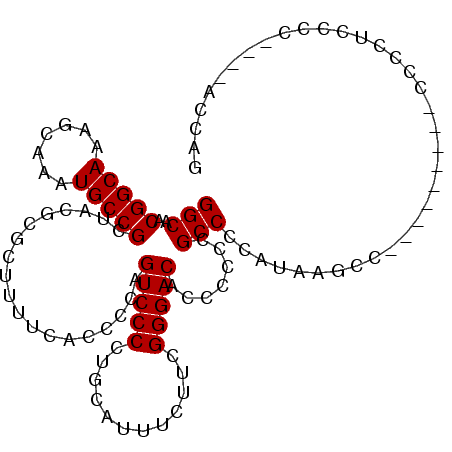
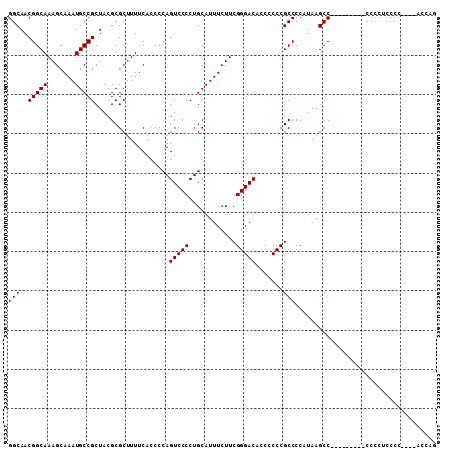
| Location | 19,153,790 – 19,153,899 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19153790 109 + 23771897 GGCAACGGCAAAGCAAAUGCCGCUACGCGCUUUUCAACCCAGUCCCCUGCAUUUCUUCGGGACACCGCCCGCCCCAUAAGCCACCUCAUCCCCUCUCCCCCCCCUCAUC (((..(((((.......)))))...................(((((............)))))................)))........................... ( -19.50) >DroSec_CAF1 16669 97 + 1 GGCAACGGCAAAGCAAAUGCCGCUACGCGCUUUUCACCCCAGUCCCCUGCAUUUCUUCGGGACACCCCCCGCCCCAUAAGCC---------CCCCUCCCCA---ACCAG (((..(((((.......)))))...................(((((............))))).......))).........---------..........---..... ( -18.70) >DroSim_CAF1 16332 96 + 1 GGCAACGGCAAAGCAAAUGCCGCUACGCGCUUUUCACCCCAGUCCCCUGCAUUUCUUCGGGACACCCCCCGCCCCAUAAGCC---------CCCCUCCCC----ACCAG (((..(((((.......)))))...................(((((............))))).......))).........---------.........----..... ( -18.70) >consensus GGCAACGGCAAAGCAAAUGCCGCUACGCGCUUUUCACCCCAGUCCCCUGCAUUUCUUCGGGACACCCCCCGCCCCAUAAGCC_________CCCCUCCCC____ACCAG (((..(((((.......)))))...................(((((............))))).......))).................................... (-18.70 = -18.70 + -0.00)

| Location | 19,153,790 – 19,153,899 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -32.33 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19153790 109 - 23771897 GAUGAGGGGGGGGAGAGGGGAUGAGGUGGCUUAUGGGGCGGGCGGUGUCCCGAAGAAAUGCAGGGGACUGGGUUGAAAAGCGCGUAGCGGCAUUUGCUUUGCCGUUGCC .....((.((.((((((.(((((..((.((((.(((((((.....)))))))....(((.(((....))).)))...)))).....))..))))).)))).)).)).)) ( -35.80) >DroSec_CAF1 16669 97 - 1 CUGGU---UGGGGAGGGG---------GGCUUAUGGGGCGGGGGGUGUCCCGAAGAAAUGCAGGGGACUGGGGUGAAAAGCGCGUAGCGGCAUUUGCUUUGCCGUUGCC .....---.........(---------.((((.(((((((.....)))))))......(.(((....))).).....)))).)(((((((((.......))))))))). ( -34.10) >DroSim_CAF1 16332 96 - 1 CUGGU----GGGGAGGGG---------GGCUUAUGGGGCGGGGGGUGUCCCGAAGAAAUGCAGGGGACUGGGGUGAAAAGCGCGUAGCGGCAUUUGCUUUGCCGUUGCC .....----........(---------.((((.(((((((.....)))))))......(.(((....))).).....)))).)(((((((((.......))))))))). ( -34.10) >consensus CUGGU____GGGGAGGGG_________GGCUUAUGGGGCGGGGGGUGUCCCGAAGAAAUGCAGGGGACUGGGGUGAAAAGCGCGUAGCGGCAUUUGCUUUGCCGUUGCC ............................((((.(((((((.....)))))))......(.(((....))).).....))))..(((((((((.......))))))))). (-32.33 = -32.33 + 0.00)



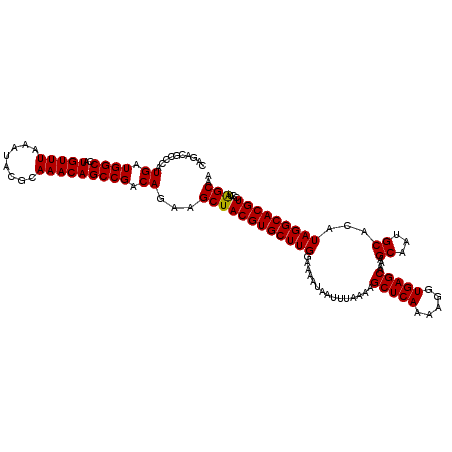
| Location | 19,153,899 – 19,154,019 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -28.76 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19153899 120 + 23771897 CAGACGCCCAUGAUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAA ..........((.((((..(((((........))))))))).))...((((((((((((...............(((((.....)))))....((...))...)))))))))...))).. ( -28.60) >DroSec_CAF1 16766 120 + 1 CAGACGCCCAUGAUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAA ..........((.((((..(((((........))))))))).))...((((((((((((...............(((((.....)))))....((...))...)))))))))...))).. ( -28.60) >DroAna_CAF1 9863 118 + 1 --UAGGUCCAUGUUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUGGCAA --........(((((((..(((((........))))))))))))...((((((((((((...............(((((.....)))))....((...))...)))))))))...))).. ( -33.20) >DroSim_CAF1 16428 120 + 1 CAGACGCCCAUGAUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAA ..........((.((((..(((((........))))))))).))...((((((((((((...............(((((.....)))))....((...))...)))))))))...))).. ( -28.60) >DroEre_CAF1 17355 117 + 1 --GACGCCCAUGAUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAA-AGCAAUGCACAUAGGCACGUCCUAGCAC --...((...((.((((..(((((........))))))))).))...))...(((((.(((.............(((((.....)))))..-.....(((......)))..))).))))) ( -28.90) >consensus CAGACGCCCAUGAUGGCCAUGUUUAAAUACGCAAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAA ..........((.((((..(((((........))))))))).))...((((((((((((...............(((((.....)))))....((...))...)))))))))...))).. (-28.76 = -28.60 + -0.16)


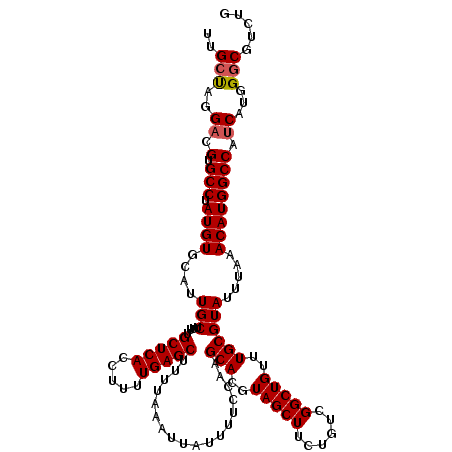
| Location | 19,153,899 – 19,154,019 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19153899 120 - 23771897 UUGCUAGGACGUGCCUAUGUGCAUUGCUUUUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAUCAUGGGCGUCUG ......((((((.(((((((((.(((.....(((((.....)))))...............)))))))))))....((..(((((((((......)))))..))))..)).).)))))). ( -31.85) >DroSec_CAF1 16766 120 - 1 UUGCUAGGACGUGCCUAUGUGCAUUGCUUUUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAUCAUGGGCGUCUG ......((((((.(((((((((.(((.....(((((.....)))))...............)))))))))))....((..(((((((((......)))))..))))..)).).)))))). ( -31.85) >DroAna_CAF1 9863 118 - 1 UUGCCAGGACGUGCCUAUGUGCAUUGCUUUUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAACAUGGACCUA-- ...(((.((((.((.(((((((.(((.....(((((.....)))))...............))))))))))))...))))(((((((((......)))))..))))....))).....-- ( -28.85) >DroSim_CAF1 16428 120 - 1 UUGCUAGGACGUGCCUAUGUGCAUUGCUUUUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAUCAUGGGCGUCUG ......((((((.(((((((((.(((.....(((((.....)))))...............)))))))))))....((..(((((((((......)))))..))))..)).).)))))). ( -31.85) >DroEre_CAF1 17355 117 - 1 GUGCUAGGACGUGCCUAUGUGCAUUGCU-UUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAUCAUGGGCGUC-- (((((.(((.((((......))))....-..(((((.....))))).............))).)))))...(((..((..(((((((((......)))))..))))..))..)))...-- ( -33.80) >consensus UUGCUAGGACGUGCCUAUGUGCAUUGCUUUUGCUCACCUUUUGAGCUUUUAAAUUAUUUUCCAAGCACGUAGCUUCUGUCGGCUGUUUGCGUAUUUAAACAUGGCCAUCAUGGGCGUCUG ..(((..((.(.(((.((((....(((....(((((.....)))))..................(((..(((((......)))))..)))))).....)))))))).))...)))..... (-29.14 = -29.38 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:47 2006