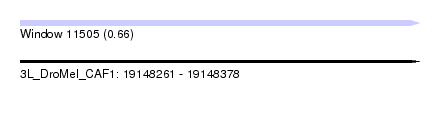
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,148,261 – 19,148,378 |
| Length | 117 |
| Max. P | 0.655292 |

| Location | 19,148,261 – 19,148,378 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -21.65 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
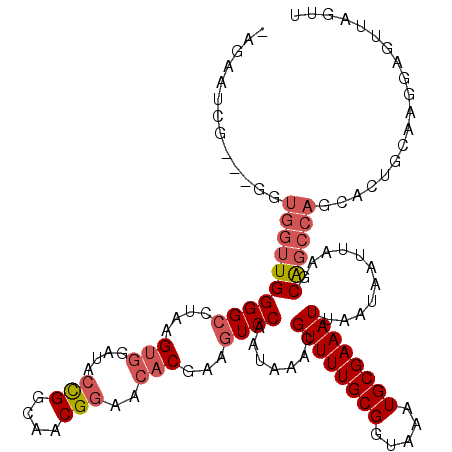
>3L_DroMel_CAF1 19148261 117 + 23771897 GAGAAUCG---GGUGGUUGGGGCCUAAGUGGAUACCGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCAGCAGUGCAAGGAGUUAGUU ........---(.((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........)))))).)................. ( -30.90) >DroSec_CAF1 11124 104 + 1 -----UCG---GGUGGUUGGGGCCUAAGUGGAUACCGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCAGCACUGCAAGG-------- -----...---(.((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........)))))).).........-------- ( -30.90) >DroAna_CAF1 3986 98 + 1 -AGAGUCCGAUGGGGGAUGGGGCCAUGG----------CGA--GCACACGAACUCCAAUAAACGUUUUGCGGCAAAUGCGAAAUUAAUAAUUAAGCGG---------AAAGUAGGAAGUU -....((((.(((((..((..((.....----------...--))...))..)))))......((((((((.....))))))))...........)))---------)............ ( -20.80) >DroSim_CAF1 10798 117 + 1 GAGAAUCG---GGUGGUUGGGGCCUAAGUGGAUACUGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCAGCACUGCAAGGAGUUAGUU ........---(.((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........)))))).)................. ( -28.70) >DroEre_CAF1 11597 111 + 1 ------UG---GGUGGUUGGGGCCUAAGUGGAUACCGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCACAACUGGAAGCAGUUAGUU ------..---.(((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........)))))))(((((....))))).... ( -37.20) >DroYak_CAF1 11727 117 + 1 GUGUAUUG---GGUGGUUGGGGCCUAAGUGGAUACCGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCACAACUGCAAGGAGUUAGUU .((((...---.(((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........)))))))...))))........... ( -34.20) >consensus _AGAAUCG___GGUGGUUGGGGCCUAAGUGGAUACCGGCAACGGAACACGAAGUCCAAUAAACGUUUUGCGGUAAAUGCGAAAUUAAUAAUUAAGCAGCCAGCACUGCAAGGAGUUAGUU .............((((((((((....(((....(((....)))..)))...)))).......((((((((.....))))))))...........))))))................... (-21.65 = -22.82 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:37 2006