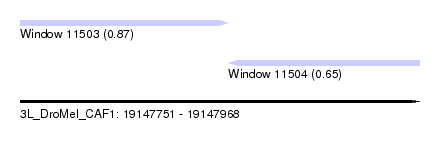
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,147,751 – 19,147,968 |
| Length | 217 |
| Max. P | 0.867839 |

| Location | 19,147,751 – 19,147,864 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.29 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -30.92 |
| Energy contribution | -31.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
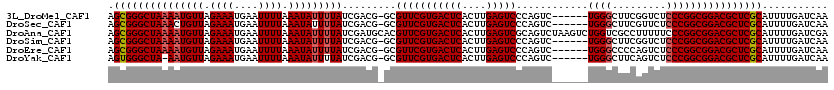
>3L_DroMel_CAF1 19147751 113 + 23771897 AGCGGGCUAAAAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG-GCGUUCGUGACUCACUUGAGUCCCAGUC------UGGGCUUCGGUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .(((((((((((((((.((((....)))).)))))))))......-..(((((((((((....)))))......------.((((....))))....))))))))))))........... ( -35.40) >DroSec_CAF1 10605 113 + 1 AGCGGGCUAAACUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG-GCGUUCGUGACUCACUUGAGUCCCAGUC------UGGGCUUCGUUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .((((((....(((((.((...((((.......))))..))))))-).(((((.(((((....))))).).(.(------((((.........))))))))))))))))........... ( -31.60) >DroAna_CAF1 3421 120 + 1 AGCGGGCUAAAAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGAUGCACGUUCGUGACUCACUUGAGUCGCAGUCUAAGUCUGGUCGCCUUUUUCCCGGCGGACGCUCGCAUUUUGAUCGA .(((((((((((((((.((((....)))).))))))))).........(((((((((((....))))))).(.(((....))).)(((........)))))))))))))........... ( -36.60) >DroSim_CAF1 10276 113 + 1 AGCGGGCUAAAAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG-GCGUUCGUGACUCACUUGAGUCCCAGUC------UGGGCUUCGGUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .(((((((((((((((.((((....)))).)))))))))......-..(((((((((((....)))))......------.((((....))))....))))))))))))........... ( -35.40) >DroEre_CAF1 11112 113 + 1 AGCGGGCUAAAAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG-GCGUUCGUGACUCACUUGAGUCCCAGUC------UGGGCCCCAGUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .(((((((((((((((.((((....)))).)))))))))......-..(((((((((((....)))))..((.(------(((...)))).))....))))))))))))........... ( -34.80) >DroYak_CAF1 11221 112 + 1 AGUGGGCUA-AAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG-GCGUUCGUGACUCACUUGAGUCCCAGUC------UGGGCUUCAGUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .((.((..(-((((((.((((....)))).)))))))..)).))(-(((((((.(((((....))))).).(.(------((((.........))))))))))))).............. ( -29.90) >consensus AGCGGGCUAAAAUGUUAGAAAUGAAUUUUAAAUAUUUUAUCGACG_GCGUUCGUGACUCACUUGAGUCCCAGUC______UGGGCUUCGGUCUCCCGGCGGACGCUCGCAUUUUGAUCAA .(((((((((((((((.((((....)))).))))))))).........(((((((((((....)))))............((((.........))))))))))))))))........... (-30.92 = -31.28 + 0.36)


| Location | 19,147,864 – 19,147,968 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
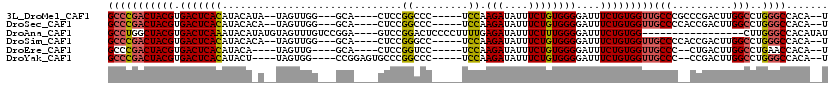
>3L_DroMel_CAF1 19147864 104 - 23771897 GCCCGACUACGUGACUCACAUACAUA--UAGUUGG---GCA----CUCCGGCCC-----UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCCGCCCGACUUGGCCUGGGCCACA--U (((((((((..((.........))..--)))))))---)).----....(((((-----.(((((.....((.(((((((((.....)))).)))))..)))))))...)))))...--. ( -40.70) >DroSec_CAF1 10718 104 - 1 GCCCGACUACGUGACUCACAUACACA--UAGUUGG---GCA----CUCCGGCCC-----UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCCCACCGACUUGGCCUGGGCCACA--U (((((((((.(((.........))).--)))))))---)).----....(((((-----.(((((.....((.((((((....(....)...)))))).)))))))...)))))...--. ( -46.60) >DroAna_CAF1 3541 99 - 1 GCCUGGCUACGUGACUCAAAUACAUAUGUAGUUUGUCCGGA----GUCCGGACUCCCCUUUUGAGAUAUUUCUUUGGGGAUUUCUGUGG-----------------CUUGGGCCACAUAU ....(((((((((...........))))))))).((((((.----..))))))(((((....((((....)))).)))))....(((((-----------------(....))))))... ( -35.20) >DroSim_CAF1 10389 104 - 1 GCCCGACUACGUGACUCACAUACACA--UAGUUGG---GCA----CUCCGGGCC-----UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCCCACCGACUUGGCCUGGGCCACA--U (((((((((.(((.........))).--)))))))---)).----.((((((((-----....(((....)))((((((....(....)...))))))......)))))))).....--. ( -43.70) >DroEre_CAF1 11225 99 - 1 GCCCGACUACGUGACUCACAUACA----UAGUUG----GCA----CUCCGGUCC-----UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCC--CUGACUUGGCCUGAACCACA--U (((.(((((.(((.......))).----))))))----)).----....(((((-----.((((((....))).))))))))..((((((((((.--.......)))...)))))))--. ( -29.60) >DroYak_CAF1 11333 103 - 1 GCCCGACUACGUGACUCACAUACU----UAGUGG----CCGGAGUGCCCGGCCC-----UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCC--CCGACUUGGCCUGGGCCACA--U (((((((((((.((..........----....((----((((.....))))))(-----(((((((....))).)))))...)))))))))(((.--.......)))..))))....--. ( -37.50) >consensus GCCCGACUACGUGACUCACAUACA_A__UAGUUGG___GCA____CUCCGGCCC_____UCCAAGAUAUUUCUGUGGGGAUUUCUGUGGUUGCCC__CCGACUUGGCCUGGGCCACA__U (((((((((((.(((((((..............................((.........)).(((....))))))))....)))))))))(((..........)))..))))....... (-16.41 = -16.63 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:36 2006