
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,147,145 – 19,147,254 |
| Length | 109 |
| Max. P | 0.955602 |

| Location | 19,147,145 – 19,147,254 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.12 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
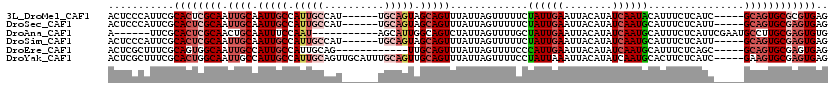
>3L_DroMel_CAF1 19147145 109 + 23771897 CUCACGCGCACUGC-----GAUGAGAAAUGUAUUGAUAUGUAAUUCAAUAGAAAAACUAAUAAACUGCUACUGCA------AUGGCAAUGGCAAUUGCAAUUGCGAGUGCGAAUGGGAGU ((((..((((((((-----(((......(.((((((........)))))).).............(((((.(((.------...))).)))))......))))).))))))..))))... ( -32.80) >DroSec_CAF1 10028 109 + 1 CUCACUCGCACUGC-----AAUGAGAAAUGCAUUGAUAUGUAAUUCAAUAGAAAAACUAAUAAACUGCUACUGCA------AUGGCAAUGGCAAUUGCAAUUGCGAGUGCGAAUGGGAGU ((((.(((((((((-----(((..(((.(((((....))))).)))...................(((((.(((.------...))).)))))......))))).))))))).))))... ( -35.40) >DroAna_CAF1 2971 103 + 1 CACACUCGCAAGGCAUUCGAAUGAGAAAUGCAUUGAUAUGUAAUUCAAUAGCAAAACUAAUAGACUGCCAAUGCU-----------AUUGGAAAUUGCAGUUGCGAGUGCGAA------U (.(((((((((((((.((.....((...((((((((........))))).)))...))....)).))))..(((.-----------..(....)..))).))))))))).)..------. ( -32.00) >DroSim_CAF1 9686 109 + 1 CUCACUCGCACUGC-----AAUGAGAAAUGCAUUGAUAUGUAAUUCAAUAGAAAAACUAAUAAACUGCUACUGCA------AUGGCAAUGGCAAUUGCAAUUGCGAGUGCGAAUGGGAGU ((((.(((((((((-----(((..(((.(((((....))))).)))...................(((((.(((.------...))).)))))......))))).))))))).))))... ( -35.40) >DroEre_CAF1 10436 103 + 1 CUCACUCGCACUGC-----GCUGAGAAAUGCAUUGAUAUGUAAUUCAAUGGGAAAACUAAUAAACUGCAA------------CUGCAAUGGCAAUGGCAAUUGCCACUGCGAAAGCGAGU ...((((((..(((-----.....(((.(((((....))))).)))..(((.....))).......))).------------.((((.(((((((....))))))).))))...)))))) ( -32.80) >DroYak_CAF1 10544 115 + 1 CUCACUCGCACUUC-----GAUGAGAAGUGCAUUGAUAUGUAAUUUAAUAGGAAAACUAAUAAACUGCAACUGCAAAUGCAACUGCAAUGGCAAUGGCAAUUGCCAGUGCGAAAGCGAGU ...(((((((((((-----.....)))))((((((...((((.((((.(((.....))).)))).))))...((((.(((...(((....)))...))).))))))))))....)))))) ( -39.20) >consensus CUCACUCGCACUGC_____AAUGAGAAAUGCAUUGAUAUGUAAUUCAAUAGAAAAACUAAUAAACUGCUACUGCA______AUGGCAAUGGCAAUUGCAAUUGCGAGUGCGAAUGGGAGU (((..((((((.............(((.(((((....))))).)))...................(((((.(((..........))).))))).((((....))))))))))....))). (-18.25 = -19.78 + 1.53)


| Location | 19,147,145 – 19,147,254 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -18.55 |
| Energy contribution | -20.55 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.44 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
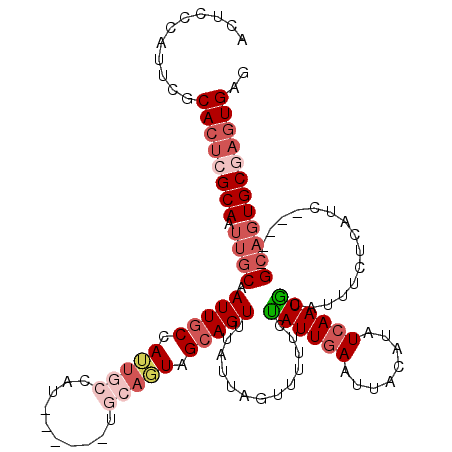
>3L_DroMel_CAF1 19147145 109 - 23771897 ACUCCCAUUCGCACUCGCAAUUGCAAUUGCCAUUGCCAU------UGCAGUAGCAGUUUAUUAGUUUUUCUAUUGAAUUACAUAUCAAUACAUUUCUCAUC-----GCAGUGCGCGUGAG .....(((.(((((((((.((((((((.((....)).))------)))))).))((((((.(((.....))).))))))......................-----).)))))).))).. ( -29.20) >DroSec_CAF1 10028 109 - 1 ACUCCCAUUCGCACUCGCAAUUGCAAUUGCCAUUGCCAU------UGCAGUAGCAGUUUAUUAGUUUUUCUAUUGAAUUACAUAUCAAUGCAUUUCUCAUU-----GCAGUGCGAGUGAG .....((((((((((.((((((((((((((.(((((...------.))))).)))))......((..(((....)))..)).......))))......)))-----)))))))))))).. ( -36.10) >DroAna_CAF1 2971 103 - 1 A------UUCGCACUCGCAACUGCAAUUUCCAAU-----------AGCAUUGGCAGUCUAUUAGUUUUGCUAUUGAAUUACAUAUCAAUGCAUUUCUCAUUCGAAUGCCUUGCGAGUGUG .------..(((((((((((.(((..........-----------.)))..((((.((.((.((...(((.(((((........))))))))...)).))..)).))))))))))))))) ( -31.10) >DroSim_CAF1 9686 109 - 1 ACUCCCAUUCGCACUCGCAAUUGCAAUUGCCAUUGCCAU------UGCAGUAGCAGUUUAUUAGUUUUUCUAUUGAAUUACAUAUCAAUGCAUUUCUCAUU-----GCAGUGCGAGUGAG .....((((((((((.((((((((((((((.(((((...------.))))).)))))......((..(((....)))..)).......))))......)))-----)))))))))))).. ( -36.10) >DroEre_CAF1 10436 103 - 1 ACUCGCUUUCGCAGUGGCAAUUGCCAUUGCCAUUGCAG------------UUGCAGUUUAUUAGUUUUCCCAUUGAAUUACAUAUCAAUGCAUUUCUCAGC-----GCAGUGCGAGUGAG .(((((((..((((((((....))))))))((((((.(------------(((.((..............((((((........)))))).....))))))-----)))))).))))))) ( -33.81) >DroYak_CAF1 10544 115 - 1 ACUCGCUUUCGCACUGGCAAUUGCCAUUGCCAUUGCAGUUGCAUUUGCAGUUGCAGUUUAUUAGUUUUCCUAUUAAAUUACAUAUCAAUGCACUUCUCAUC-----GAAGUGCGAGUGAG .(((((((..((((((.(((.(((.(((((....))))).))).)))))).)))((((((.(((.....))).))))))..........(((((((.....-----)))))))))))))) ( -39.60) >consensus ACUCCCAUUCGCACUCGCAAUUGCAAUUGCCAUUGCCAU______UGCAGUAGCAGUUUAUUAGUUUUUCUAUUGAAUUACAUAUCAAUGCAUUUCUCAUC_____GCAGUGCGAGUGAG ...........((((((((.((((.(((((.(((((..........))))).))))).............((((((........))))))................)))))))))))).. (-18.55 = -20.55 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:34 2006