| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,145,001 – 19,145,222 |
| Length | 221 |
| Max. P | 0.641300 |

| Location | 19,145,001 – 19,145,119 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19145001 118 + 23771897 -AACAGGACGUAGAAAAGGAGUCUCGACUCCUGA-GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGCUUAAUGCCAUCCACCGUUAACAAAUUGUUUACCCA -..(((..(((((((.((((((....))))))..-..)))))))..))).............(..((((((((....(((((.((.....)).)))))..(....)..))))))))..). ( -33.10) >DroSec_CAF1 7900 118 + 1 -AAAAGGACGUAGAAAAGGAGUCUCGACUCCUGA-GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGCUUAAUGCCAUCCACCGUUGACAAAUUGUUUACCCA -..((((.(((((((.((((((....))))))..-..)))))))))))((((.....)))).(..((((((((....(((((.((.....)).)))))..(....)..))))))))..). ( -33.20) >DroAna_CAF1 1015 120 + 1 UGCCAGGACACAGGAAGGAUUUCUAAACAGCUGAAGGUUGAGCGCUUUAGAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGUUUAAUGCCAUCCACUGUUGACAAAUUGUUUACCCA .....((..((((((((((((((((((..(((........)))..)))))))))).)))))))).((((((((....(((((.((.....)).))))).((....)).)))))))).)). ( -32.70) >DroSim_CAF1 7553 118 + 1 -AACAGGACGUAGAAAAGGAGUCUCGACUCCUGA-GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGCUUAAUGCCAUCCACCGUUGACAAAUUGUUUACCCA -..(((..(((((((.((((((....))))))..-..)))))))..))).............(..((((((((....(((((.((.....)).)))))..(....)..))))))))..). ( -33.10) >DroEre_CAF1 8321 118 + 1 -AGCAGGACGUAGAAAAGGAGUUUCGACUCCUGA-GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGCUUAAUGCCAUCCACCGUUGACAAAUUGUUUACCCA -.(((((((.(.....((((((....)))))).)-.))))))).....((((.....)))).(..((((((((....(((((.((.....)).)))))..(....)..))))))))..). ( -32.30) >DroYak_CAF1 8349 118 + 1 -AGCAGGACGUAGAAAAGGAGUUUCGACUCCUGA-GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAACAUUAUGGAUCGCUUAAUGCCAUCUACCGUUGACAAAUUGUUUACCCA -.(((((((.(.....((((((....)))))).)-.))))))).....((((.....)))).(..(((((((.....(((((.((.....)).)))))..(....)...)))))))..). ( -28.70) >consensus _AACAGGACGUAGAAAAGGAGUCUCGACUCCUGA_GGUUCUGCGCUUUGAAAAUUACUUUCUGUUUAGACAAUAUUAUGGAUCGCUUAAUGCCAUCCACCGUUGACAAAUUGUUUACCCA ....(((.(((((((.((((((....)))))).....))))))))))...............(..((((((((....(((((.((.....)).)))))..(....)..))))))))..). (-25.80 = -25.83 + 0.03)


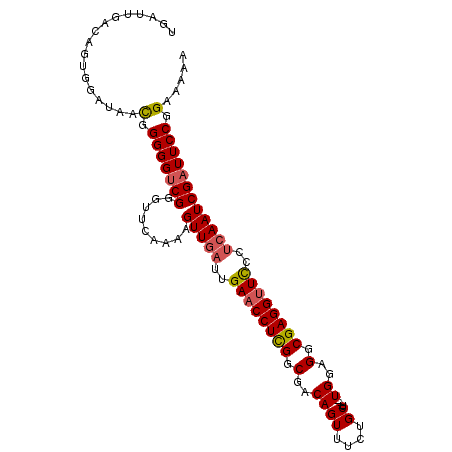
| Location | 19,145,119 – 19,145,222 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19145119 103 - 23771897 UGAUUGACAGUGGUUAACGGGGGACGGGUUCAAAAGUUGAUUGAACCUCGGCGACAGUUUCUGCUUUUUGGAGGCGAGGUUCCCUCAAUCGAUUCCGGAAAAA .((((((..((.....)).((((((.(((((((.......)))))))(((.(..(((..........)))..).))).))))))))))))............. ( -29.60) >DroSec_CAF1 8018 100 - 1 UGAUUGACAGUGAAUAACGGGGGUCGGGUUUAAAAGUUGAUUGAACCUCGGCGACAGUUUCUGCU---UGGAGGCGAGGUUCCCUCAAUCGAUUCCGGAAAAA .................(.(((((((.........(((((..((((((((.(..((((....)).---))..).))))))))..)))))))))))).)..... ( -31.80) >DroSim_CAF1 7671 100 - 1 UGAUUGACAGUGGAUAACAGGGGUCGGGUUCAAAAGUUUAUUGAACCUCGGCGACAGUUUCUGCU---UGCAGGCGAGGCUCCCACAAUCGAUUCCGGAAAAA .((((((..((((......(..(((((((((((.......))))))).))))..)((((((.(((---....))))))))).))))..))))))......... ( -31.90) >DroEre_CAF1 8439 100 - 1 UGAUUGACAAAGGAAAACGGGGGUCGUGCUCAAAAGUUGAUUGAACCUUGGCGACAGUUUCUGCU---UGGAGGCGAGGUUCCCUCAAUCGAUUCCGGAAAAA .................(.(((((((.........(((((..((((((((.(..((((....)).---))..).))))))))..)))))))))))).)..... ( -29.70) >DroYak_CAF1 8467 100 - 1 UGAUUGACAGAGGAAAAUGGGGGUCGGGUUCAAAAGUUGAUUGAACCUCGGCGACAGUUUCAGCU---UGGAGGCGAGGUUUCCUCAAUCGAUUCCGGAAAAA .((((((..(((((((...(..(((((((((((.......))))))).))))..).(((((....---..)))))....)))))))..))))))......... ( -32.00) >consensus UGAUUGACAGUGGAUAACGGGGGUCGGGUUCAAAAGUUGAUUGAACCUCGGCGACAGUUUCUGCU___UGGAGGCGAGGUUCCCUCAAUCGAUUCCGGAAAAA .................(.(((((((.........(((((..((((((((.(..((((....))....))..).))))))))..)))))))))))).)..... (-24.82 = -25.14 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:28 2006