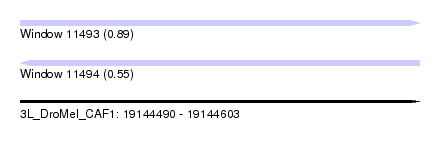
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,144,490 – 19,144,603 |
| Length | 113 |
| Max. P | 0.890883 |

| Location | 19,144,490 – 19,144,603 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.34 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -26.03 |
| Energy contribution | -26.95 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19144490 113 + 23771897 ----GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUC---AAAAGGACACAAAAGAAAGCAAAGGACACACAUGCACCAGAAC ----..(((((...(((.((((.((((......))))))))..)))...(((..(((.(((((.((..(((((.---.............)))))))..))))))))..)))..))))). ( -31.14) >DroSec_CAF1 7383 113 + 1 ----GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUC---AAAAGGACACAAAAGAAAGCAAAGGACACACAUGCAUCAGAAU ----..(((((...(((.((((.((((......))))))))..)))...(((..(((.(((((.((..(((((.---.............)))))))..))))))))..)))..))))). ( -31.04) >DroAna_CAF1 691 117 + 1 CUCUGUCUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUCGAAAAAAGGACACAAAAGAAACCAAAGGACACACAUGCACCGG--- ...(((((((((..(((.((((.((((......))))))))..)))...)))))(((.(((((....((((....)))).)))))))).............))))............--- ( -28.30) >DroSim_CAF1 7001 113 + 1 ----GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUC---AAAAGGACACAAAAGAAAGCAAAGGACACACAUGCACCAGAAC ----..(((((...(((.((((.((((......))))))))..)))...(((..(((.(((((.((..(((((.---.............)))))))..))))))))..)))..))))). ( -31.14) >DroEre_CAF1 7888 113 + 1 ----GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGCCGUUUUCUC---AAAAGGACACAAAAGAAACCAAACGACACACAUGCACCAGAAC ----..(((((....(((((((.((((......)))))))).(((....)))..(((.(((((...........---...)))))))).....................)))..))))). ( -25.74) >DroYak_CAF1 7900 108 + 1 ----GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUC---AAAAGGACACAAAAGAAACCAAAGGACACGCAUGCACC----- ----...(((((..(((.((((.((((......))))))))..)))...))))).((.(((((.....(((((.---.............)))))....))))))).........----- ( -25.74) >consensus ____GUUUCUGUCUGGCAGCUGUGAAAAUCCAAUUUCCAGCACGCCUAAGCGGAUGUAGUCCUGGCAUUUUCUC___AAAAGGACACAAAAGAAACCAAAGGACACACAUGCACCAGAAC ......(((((...(((.((((.((((......))))))))..)))...(((..(((.(((((.....(((((.................)))))....))))))))..)))..))))). (-26.03 = -26.95 + 0.92)

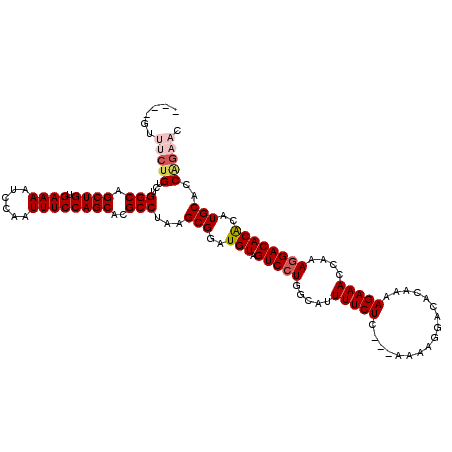
| Location | 19,144,490 – 19,144,603 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.34 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -29.48 |
| Energy contribution | -31.15 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.546041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19144490 113 - 23771897 GUUCUGGUGCAUGUGUGUCCUUUGCUUUCUUUUGUGUCCUUUU---GAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC---- .((((((((.(((((.(((((..((((((((..(......)..---))))))..)).)))))))))).)))...((((.((((((((......)))).))))))))...)))))..---- ( -38.50) >DroSec_CAF1 7383 113 - 1 AUUCUGAUGCAUGUGUGUCCUUUGCUUUCUUUUGUGUCCUUUU---GAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC---- .(((((..((..(((((((((..((((((((..(......)..---))))))..)).))))).))))..))...((((.((((((((......)))).))))))))...)))))..---- ( -36.20) >DroAna_CAF1 691 117 - 1 ---CCGGUGCAUGUGUGUCCUUUGGUUUCUUUUGUGUCCUUUUUUCGAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAGACAGAG ---..((((.(((((.(((((..(((...(((((...........)))))....))))))))))))).))))..((((.((((((((......)))).)))))))).............. ( -33.40) >DroSim_CAF1 7001 113 - 1 GUUCUGGUGCAUGUGUGUCCUUUGCUUUCUUUUGUGUCCUUUU---GAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC---- .((((((((.(((((.(((((..((((((((..(......)..---))))))..)).)))))))))).)))...((((.((((((((......)))).))))))))...)))))..---- ( -38.50) >DroEre_CAF1 7888 113 - 1 GUUCUGGUGCAUGUGUGUCGUUUGGUUUCUUUUGUGUCCUUUU---GAGAAAACGGCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC---- ..((((((((.((((....(((..((((((..((((((((.((---(......))).))))).)))..(((....)))....))))))..)))...)))))).)))))).......---- ( -35.80) >DroYak_CAF1 7900 108 - 1 -----GGUGCAUGCGUGUCCUUUGGUUUCUUUUGUGUCCUUUU---GAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC---- -----((((.(((...(((((..((((((((..(......)..---)))))...))))))))..))).))))..((((.((((((((......)))).))))))))..........---- ( -31.00) >consensus GUUCUGGUGCAUGUGUGUCCUUUGCUUUCUUUUGUGUCCUUUU___GAGAAAAUGCCAGGACUACAUCCGCUUAGGCGUGCUGGAAAUUGGAUUUUCACAGCUGCCAGACAGAAAC____ .((((((((.(((((.(((((....((((((..(........)...)))))).....)))))))))).)))...((((.((((((((......)))).))))))))...)))))...... (-29.48 = -31.15 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:26 2006