| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,143,870 – 19,143,967 |
| Length | 97 |
| Max. P | 0.802464 |

| Location | 19,143,870 – 19,143,967 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -13.83 |
| Energy contribution | -17.33 |
| Covariance contribution | 3.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
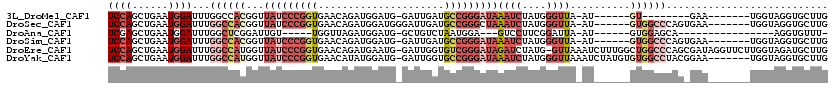
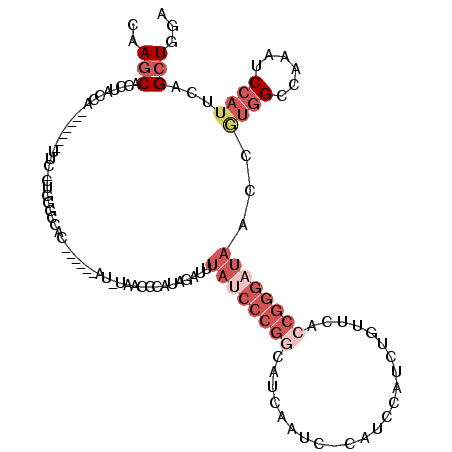
>3L_DroMel_CAF1 19143870 97 + 23771897 UCCAGCUGAAUGGAUUUGGCCACGGUUAUCCCGGUGAACAGAUGGAUG-GAUUGAUGCCGGGAUAAAUCUAUGGGUUA-AU------GU--------GAA-------UGGUAGGUGCUUG ((((......)))).((((((.((.(((((((((((....(((.....-.)))..))))))))))).....)))))))-).------..--------...-------............. ( -24.40) >DroSec_CAF1 6754 106 + 1 UCCAGCUGAAUGGAUUUGGCCACGGUUAUCCCGGUGAACAGAUGGAUGGGAUUGAUGCCGGGCUAAAUCUAUGGGUUA-AU------GUGGCCCAGUGAA-------UGGUAGGUGCUUG .((.(((..((((((((((((.((((.((((((.............))))))....))))))))))))))))(((((.-..------..)))))......-------.))).))...... ( -39.32) >DroAna_CAF1 177 87 + 1 UCGAGCUGAAUGGAUUUGGCUCGGAUUGU-----UGGUUAGAUGGAUG-GCUGUCUAAUGGA---GUCCUUCGGAUUA-AU------GUGGAGCA----------------AGGUGUUU- ((((((..((....))..)))))).((((-----(.(((((((((...-.)))))))))..(---((((...))))).-..------....))))----------------).......- ( -24.10) >DroSim_CAF1 6345 105 + 1 UCCAGCUGAAUGGAUUUGGCCACGGUUAUCCCGGUGAACAGAUGGAUG-GAUUGAUGCCGGGAUAAAUCUAUGGGUUA-AU------GUGGCCCAGUGAA-------UGGUAGGUGCUUG ..((.(((.((..(((.(((((((..((((((((((....(((.....-.)))..))))))))))((((....)))).-.)------)))))).)))..)-------)..))).)).... ( -32.30) >DroEre_CAF1 7245 118 + 1 UCCAGCUGAAUGGAUUUGGCCAUGGUUAUCCCGGUGAACAGAUGAAUG-GAUUGGUGUCGGGAUAGAUCUAUG-GUUAAAUCUUUGGCUGGCCCAGCGAUAGGUUCUUGGUAGAUGCUUG .(((((..(..((((((((((((((((((((((..(....(((.....-.)))..)..))))))))..)))))-))))))))))..))))).((((.((.....)))))).......... ( -44.00) >DroYak_CAF1 7250 112 + 1 UCCAGCUGAAUGGAUUUGGCCAUGGUUAUCCCGGUGAACAUAUGGAUG-GAUUGGUGCCGGGAUAAAUCUAUGGGUUAAAUCUAUGUGUGGCCUACGGAA-------UGGUAGGUGCUUG ...(((...(((((((((((((((((((((((((((..(((....)))-......)))))))))))..)))).)))))))))))).....((((((....-------..))))))))).. ( -42.30) >consensus UCCAGCUGAAUGGAUUUGGCCACGGUUAUCCCGGUGAACAGAUGGAUG_GAUUGAUGCCGGGAUAAAUCUAUGGGUUA_AU______GUGGCCCAG_GAA_______UGGUAGGUGCUUG ((((......))))...((((((...(((((((((.....................)))))))))((((....))))..........))))))........................... (-13.83 = -17.33 + 3.50)
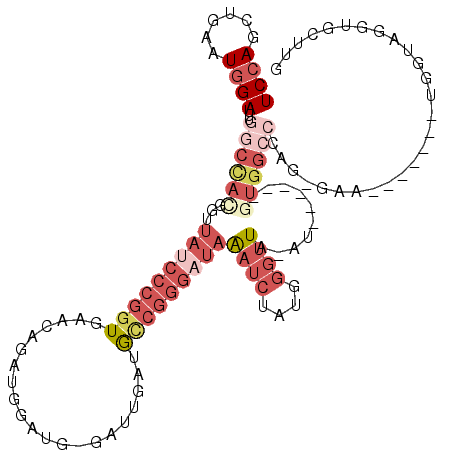
| Location | 19,143,870 – 19,143,967 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -7.38 |
| Energy contribution | -9.55 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19143870 97 - 23771897 CAAGCACCUACCA-------UUC--------AC------AU-UAACCCAUAGAUUUAUCCCGGCAUCAAUC-CAUCCAUCUGUUCACCGGGAUAACCGUGGCCAAAUCCAUUCAGCUGGA .............-------...--------..------..-....(((..(..(((((((((...((...-........))....))))))))))..))).....((((......)))) ( -17.80) >DroSec_CAF1 6754 106 - 1 CAAGCACCUACCA-------UUCACUGGGCCAC------AU-UAACCCAUAGAUUUAGCCCGGCAUCAAUCCCAUCCAUCUGUUCACCGGGAUAACCGUGGCCAAAUCCAUUCAGCUGGA ..(((........-------.....((((....------..-...))))..(((((.((((((.....(((((...............)))))..))).))).)))))......)))... ( -24.26) >DroAna_CAF1 177 87 - 1 -AAACACCU----------------UGCUCCAC------AU-UAAUCCGAAGGAC---UCCAUUAGACAGC-CAUCCAUCUAACCA-----ACAAUCCGAGCCAAAUCCAUUCAGCUCGA -.....(((----------------(.......------..-.......))))..---....(((((....-......)))))...-----......(((((............))))). ( -8.19) >DroSim_CAF1 6345 105 - 1 CAAGCACCUACCA-------UUCACUGGGCCAC------AU-UAACCCAUAGAUUUAUCCCGGCAUCAAUC-CAUCCAUCUGUUCACCGGGAUAACCGUGGCCAAAUCCAUUCAGCUGGA .............-------.......((((((------.(-((.....)))..(((((((((...((...-........))....)))))))))..))))))...((((......)))) ( -25.30) >DroEre_CAF1 7245 118 - 1 CAAGCAUCUACCAAGAACCUAUCGCUGGGCCAGCCAAAGAUUUAAC-CAUAGAUCUAUCCCGACACCAAUC-CAUUCAUCUGUUCACCGGGAUAACCAUGGCCAAAUCCAUUCAGCUGGA ..........(((.((.....))(((((((((......((((((..-..))))))(((((((.........-...............)))))))....))))).........))))))). ( -25.26) >DroYak_CAF1 7250 112 - 1 CAAGCACCUACCA-------UUCCGUAGGCCACACAUAGAUUUAACCCAUAGAUUUAUCCCGGCACCAAUC-CAUCCAUAUGUUCACCGGGAUAACCAUGGCCAAAUCCAUUCAGCUGGA ..(((.(((((..-------....))))).........(((((...((((.(..(((((((((........-..............)))))))))).))))..)))))......)))... ( -25.75) >consensus CAAGCACCUACCA_______UUC_CUGGGCCAC______AU_UAACCCAUAGAUUUAUCCCGGCAUCAAUC_CAUCCAUCUGUUCACCGGGAUAACCGUGGCCAAAUCCAUUCAGCUGGA ..(((..................................................((((((((.......................))))))))...((((......))))...)))... ( -7.38 = -9.55 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:23 2006