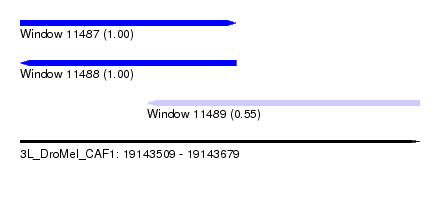
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,143,509 – 19,143,679 |
| Length | 170 |
| Max. P | 0.999921 |

| Location | 19,143,509 – 19,143,601 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.44 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19143509 92 + 23771897 AAAGA-----GAAAUCGGUGGCACUAUAUCGC-CUAUAUA----CAUACAUAUAUACGAAUACA------------------GUAUGUGUGUGUGAAGCAAUCACACCAAAUAAAUCAUU ...((-----....))((((((........))-)......----(((((((((((((.......------------------)))))))))))))..........)))............ ( -25.10) >DroSec_CAF1 6419 74 + 1 AAAGA-----GAAAUCGGUGGCACUAUACCGUGCCAUGUG----UGUACA-------------------------------------UGUGUGUGAAGCAAUCACACCAAAUAAAUCAUU ...((-----....)).(((((((......)))))))(((----..((..-------------------------------------((.((((((.....))))))))..))...))). ( -21.10) >DroSim_CAF1 5988 92 + 1 AAAGA-----GAAAUCGGUGGCACUAUACCGC-CUAUAUA----CAUACAUAUAUACGAAUAUA------------------CUAUGUGUGUGUGAAGCAAUCACACCAAAUAAAUCAUU ...((-----....))((((((........))-((.((((----((((((((((((....))))------------------.)))))))))))).))......))))............ ( -22.10) >DroEre_CAF1 6861 111 + 1 AAAGCGCAAUGGAAUCGGUGGCAGUAUAUCGC-CUAUAU--------ACAAAUAUACGAAUAUACUUGUAGUACACACAUACAUAUGUGUGUGUGAAGCAAUCACACCAAAUAAAUCAUU ................(((((((((((((((.-.(((((--------....))))))).)))))).)))....(((((((((....))))))))).........))))............ ( -27.80) >DroYak_CAF1 6888 96 + 1 AAAGA-----GGAAUCGGCGGCAGUAUAUCGC-CUAUAUAUGUACAUACAUAUAUACGAAUAUA------------------GUAUGUGUGUGUGAAGUAAUCACACCAAAUAAAUCAUU .....-----((....(((((.......))))-)..........(((((((((((((.......------------------)))))))))))))...........))............ ( -24.60) >consensus AAAGA_____GAAAUCGGUGGCACUAUAUCGC_CUAUAUA____CAUACAUAUAUACGAAUAUA__________________GUAUGUGUGUGUGAAGCAAUCACACCAAAUAAAUCAUU ...............(((((......)))))........................................................((.((((((.....))))))))........... (-10.70 = -10.66 + -0.04)

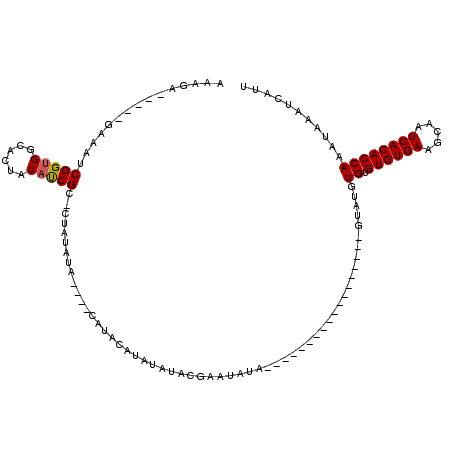
| Location | 19,143,509 – 19,143,601 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -10.27 |
| Energy contribution | -11.48 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19143509 92 - 23771897 AAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAC------------------UGUAUUCGUAUAUAUGUAUG----UAUAUAG-GCGAUAUAGUGCCACCGAUUUC-----UCUUU ...........((((((((.....)))))).))..(((------------------(((((((.(((((((....)----)))))).-).)))))))))...........-----..... ( -21.30) >DroSec_CAF1 6419 74 - 1 AAUGAUUUAUUUGGUGUGAUUGCUUCACACACA-------------------------------------UGUACA----CACAUGGCACGGUAUAGUGCCACCGAUUUC-----UCUUU .......(((.((((((((.....)))))).))-------------------------------------.)))..----....((((((......))))))........-----..... ( -19.50) >DroSim_CAF1 5988 92 - 1 AAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAG------------------UAUAUUCGUAUAUAUGUAUG----UAUAUAG-GCGGUAUAGUGCCACCGAUUUC-----UCUUU ......((((.((((((((.....)))))).)).))))------------------(((((.(((((....)))))----.)))))(-(.(((.....))).))......-----..... ( -20.60) >DroEre_CAF1 6861 111 - 1 AAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAUGUAUGUGUGUACUACAAGUAUAUUCGUAUAUUUGU--------AUAUAG-GCGAUAUACUGCCACCGAUUCCAUUGCGCUUU ............((((..((.(..((..((((((((....))))))))..((((((((((....)))))))))--------)....(-(((......))))...))..).))..)))).. ( -30.20) >DroYak_CAF1 6888 96 - 1 AAUGAUUUAUUUGGUGUGAUUACUUCACACACACAUAC------------------UAUAUUCGUAUAUAUGUAUGUACAUAUAUAG-GCGAUAUACUGCCGCCGAUUCC-----UCUUU .(((.........((((((.....))))))..((((((------------------(((((....))))).)))))).))).....(-(((.........))))......-----..... ( -22.70) >consensus AAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAC__________________UAUAUUCGUAUAUAUGUAUG____UAUAUAG_GCGAUAUAGUGCCACCGAUUUC_____UCUUU ..........(((((((((.....)))).................................((((...((((((......))))))..)))).........))))).............. (-10.27 = -11.48 + 1.21)

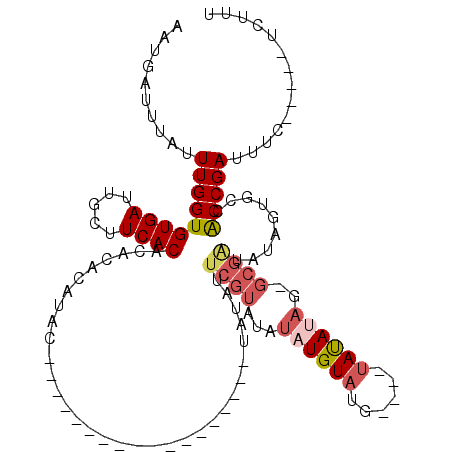
| Location | 19,143,563 – 19,143,679 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19143563 116 - 23771897 UCUCGCCACUGCAUCCAUCUCUUUCUCUUCGCAUACGACCAUCUUC--CUCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAC-- ......((.(((((((((.((.......(((((...((........--.))..)))))....)).))))...))))).)).......(((.((((((((.....)))))).)).))).-- ( -21.70) >DroSec_CAF1 6460 109 - 1 UCUCGCCACUGCAUCCGCCUCUUUCUCUUUGCAUACGACCAUCUC----UCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUGCUUCACACACA------- ......((.(((((..(((..((((...(((((...((.......----))..)))))....)))).)))..))))).)).............((((((.....))))))...------- ( -23.70) >DroSim_CAF1 6042 118 - 1 UCUCGCCACUGCAUCCGCCUCUUUCUCUUUGCAUACGACCAUCUCUCUCUCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAG-- ......((.(((((..(((..((((...(((((....................)))))....)))).)))..))))).)).......(((.((((((((.....)))))).)).))).-- ( -24.15) >DroEre_CAF1 6932 116 - 1 UCUCGCCCCUGCAUCCGUCUCUUUCCCUUUGCAUACGACCAUCUC----UCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAUGU .........(((((..(((..((((...(((((...((.......----))..)))))....)))).)))..)))))..........(((.((((((((.....)))))).)).)))... ( -20.30) >DroYak_CAF1 6946 114 - 1 UCUCGCCCCUGCAUCCGUCCCUUUCUCUUUGCAUACGACCGUCUC----UCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUACUUCACACACACAUAC-- .........(((((..(.((.((((...(((((...((.......----))..)))))....)))).)))..)))))..........(((.((((((((.....)))))).)).))).-- ( -20.50) >consensus UCUCGCCACUGCAUCCGUCUCUUUCUCUUUGCAUACGACCAUCUC____UCUCUGCGACUCCGAAAUGGCAAAUGCAUUGAAUGAUUUAUUUGGUGUGAUUGCUUCACACACACAUAC__ ......((.(((((..(((..((((...(((((....................)))))....)))).)))..))))).)).............((((((.....)))))).......... (-19.91 = -20.11 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:21 2006