
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
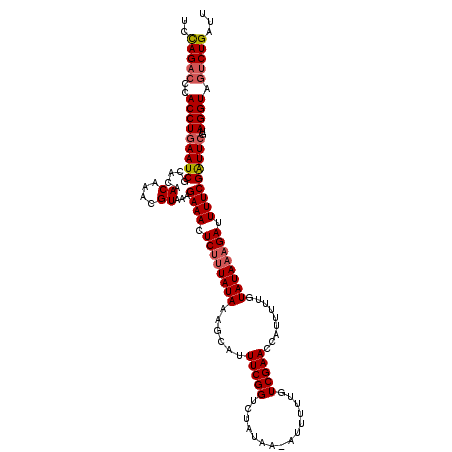
| Location | 2,123,578 – 2,123,698 |
| Length | 120 |
| Max. P | 0.804789 |

| Location | 2,123,578 – 2,123,698 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
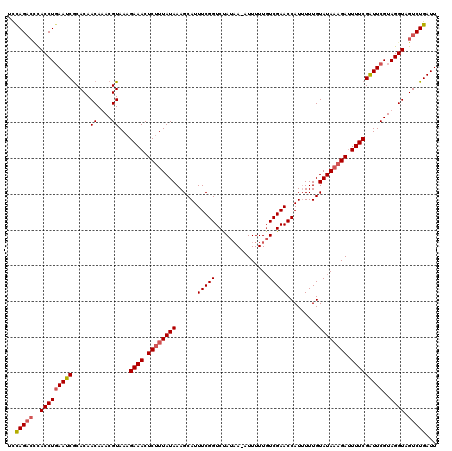
>3L_DroMel_CAF1 2123578 120 + 23771897 UCCAGACCCACCUGAAUCGCACAACAAACGUAAAGAAACUCUUUAUAAAGCAUUUCGGUCUAUAAGUUUUUUGUCGAACCAUGUUUGUAUAAAGAUUUUCGAUUCGUAGGUAGUCUGAUU ..(((((..(((((((((.....((....))...((((.(((((((((((((((((((.(............))))))..)))))).)))))))).)))))))))..)))).)))))... ( -30.90) >DroSec_CAF1 175823 119 + 1 UCCAGACCCACCUGAAUCGCACAACAAACGUAAAGAAACUCUUUAUAAAGCAUUUCGGUCUAUAA-UUUUUUGUCGAACCAUUUUUGUAUAAAGAUUUUCGAUUCGUAGGUAGUCUGAUU ..(((((..(((((((((.....((....))...((((.((((((((.........((((.((((-....)))).).))).......)))))))).)))))))))..)))).)))))... ( -27.39) >DroEre_CAF1 180514 119 + 1 ACUAGACUCACCUCAAUCGCACAACAUACGUAAAGAAACUCUUUAUAAAGCAUUUCGGUCCAUCA-AUUUUUGUCGAACCAUUUUUGUAUAAAGAUUUUCGGUUCGUAGGUAGUCUGAUU ..((((((.(((((((((.....((....))...((((.((((((((.........(((.(..((-.....))..).))).......)))))))).)))))))).).))))))))))... ( -23.99) >DroYak_CAF1 173426 115 + 1 -UUAG---UACCUGAAUCACACAACAUACGUAAAGAAACUCCUUAUAAAGCAUUUCGGUCUACAA-AUUUUUCUCGAACCAUUUUUGUAUACAGAAUUUCGAUUCGUAGGUAGUCUGAUU -....---......(((((.((.((.((((....((((.((..(((((((...(((((.......-.......)))))....)))))))....)).))))....)))).)).)).))))) ( -16.84) >consensus UCCAGACCCACCUGAAUCGCACAACAAACGUAAAGAAACUCUUUAUAAAGCAUUUCGGUCUAUAA_AUUUUUGUCGAACCAUUUUUGUAUAAAGAUUUUCGAUUCGUAGGUAGUCUGAUU ..(((((..(((((((((.....((....))...((((.((((((((......(((((...............))))).........)))))))).)))))))))..)))).)))))... (-18.93 = -19.75 + 0.81)

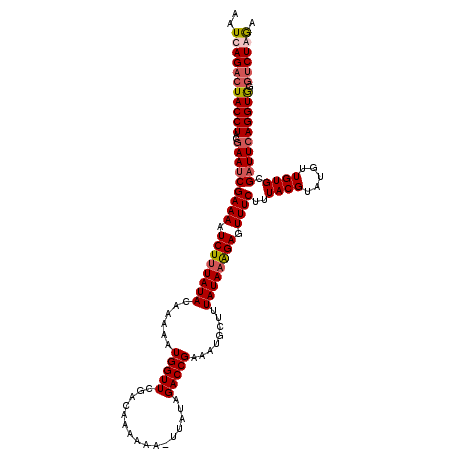
| Location | 2,123,578 – 2,123,698 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -22.57 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
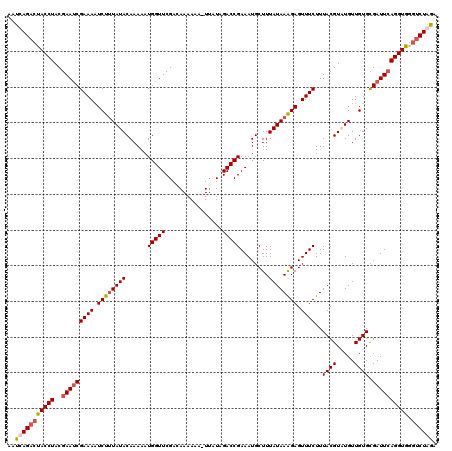
>3L_DroMel_CAF1 2123578 120 - 23771897 AAUCAGACUACCUACGAAUCGAAAAUCUUUAUACAAACAUGGUUCGACAAAAAACUUAUAGACCGAAAUGCUUUAUAAAGAGUUUCUUUACGUUUGUUGUGCGAUUCAGGUGGGUCUGGA ..(((((((((((..((((((...........((((((..((((................))))(((((.(((....))).))))).....))))))....)))))))))).))))))). ( -29.95) >DroSec_CAF1 175823 119 - 1 AAUCAGACUACCUACGAAUCGAAAAUCUUUAUACAAAAAUGGUUCGACAAAAAA-UUAUAGACCGAAAUGCUUUAUAAAGAGUUUCUUUACGUUUGUUGUGCGAUUCAGGUGGGUCUGGA ..(((((((((((..(((((((((.((((((((......(((((..........-.....)))))........)))))))).))))..((((.....)))).))))))))).))))))). ( -29.20) >DroEre_CAF1 180514 119 - 1 AAUCAGACUACCUACGAACCGAAAAUCUUUAUACAAAAAUGGUUCGACAAAAAU-UGAUGGACCGAAAUGCUUUAUAAAGAGUUUCUUUACGUAUGUUGUGCGAUUGAGGUGAGUCUAGU ....(((((((((.(((...((((.((((((((......(((((((.((.....-)).)))))))........)))))))).))))....(((((...))))).))))))).)))))... ( -29.94) >DroYak_CAF1 173426 115 - 1 AAUCAGACUACCUACGAAUCGAAAUUCUGUAUACAAAAAUGGUUCGAGAAAAAU-UUGUAGACCGAAAUGCUUUAUAAGGAGUUUCUUUACGUAUGUUGUGUGAUUCAGGUA---CUAA- (((((.((.((.((((.(..((((((((.((((......(((((((((.....)-)))..)))))........)))).))))))))..).)))).)).)).)))))......---....- ( -22.34) >consensus AAUCAGACUACCUACGAAUCGAAAAUCUUUAUACAAAAAUGGUUCGACAAAAAA_UUAUAGACCGAAAUGCUUUAUAAAGAGUUUCUUUACGUAUGUUGUGCGAUUCAGGUGGGUCUAGA ..(((((((((((..(((((((((.((((((((......(((((................)))))........)))))))).))))..((((.....)))).)))))))))).)))))). (-22.57 = -23.75 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:05 2006