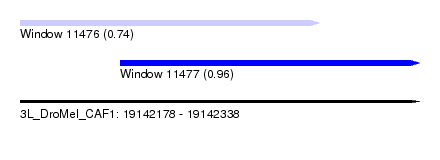
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,142,178 – 19,142,338 |
| Length | 160 |
| Max. P | 0.958980 |

| Location | 19,142,178 – 19,142,298 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19142178 120 + 23771897 UUGGGAAACGCAGUUAGAGUCAGACAAUAACUGGUCAAGGUCAAGCAGAACGGAAAACCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUU ..(((.((((((....((.((((.......))))))(((((((.((((..(((....))).(((((........)))))..)).))....))))))))))))).)))............. ( -28.90) >DroSec_CAF1 5104 120 + 1 UUGGGAAACGCAGUUAGAGUCAGACAAUAACAGGUCAAGGUCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUU ..(((.((((((..........(((........)))(((((((....((..(..(((((...............)))))..)...))...))))))))))))).)))............. ( -28.86) >DroSim_CAF1 4670 120 + 1 UUGCGAAACGCAGUUAGAGUCAGACAAUAACUGGUCAAGGUCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUU (((((...)))))((((((((((.......)))((.(((((((....((..(..(((((...............)))))..)...))...))))))).))...))))))).......... ( -28.96) >DroEre_CAF1 5522 120 + 1 UUGGGAAACGCAGUUAGAGUCAGACAAUAACUGGUCAAGGUCAAGUAGAACGGAAACCCGAAAUCCAUGCCAAUGGGUUAACUAGUCCAGUGACCUUUGCGUUGCUCUAAAAUGAACUUU ..(((.((((((((((..(.....)..)))))(((((.((.(.(((....(((....))).(((((((....))))))).))).).))..)))))...))))).)))............. ( -33.90) >DroYak_CAF1 5447 120 + 1 UUGGGAAACGCAGUUAGAGUCGGACAAUAACUGGUCAAGGUCAAGUAGAACGGAAACCCGAAAUCCGUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUGCUCUAAAAUGAACUUU ..(((.((((((((((..(.....)..)))))(((((..(.(.(((....(((....))).(((((........))))).))).).)...)))))...))))).)))............. ( -29.80) >consensus UUGGGAAACGCAGUUAGAGUCAGACAAUAACUGGUCAAGGUCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUU ..(((.((((((..........(((........)))(((((((....((.(((....))).(((((........)))))......))...))))))))))))).)))............. (-27.66 = -27.86 + 0.20)


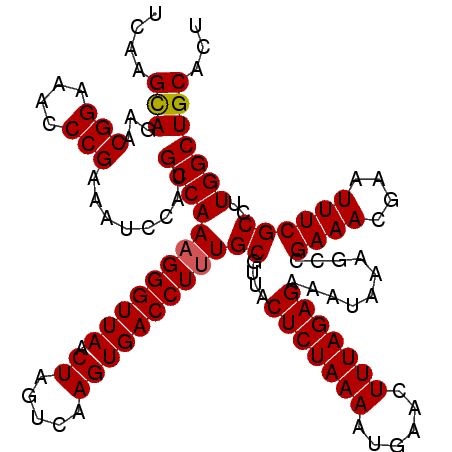
| Location | 19,142,218 – 19,142,338 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19142218 120 + 23771897 UCAAGCAGAACGGAAAACCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUUAGAGAAAUAAAACCGAAACGAAUUUCGCCUUGGCUGCACU ....((((..(((....)))......((((..((((((((.((.....))))))))))))))..(((((((......)))))))........((((..((.....))..))))))))... ( -28.30) >DroSec_CAF1 5144 120 + 1 UCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUUAGAGGCAUACAUCCGAAACGAAUUUCGCCUUGGCUGCACU ....((((..(((....)))......(((((.((((((((.((.....)))))))))).......((((((......)))))))))))....((((..((.....))..))))))))... ( -30.80) >DroSim_CAF1 4710 120 + 1 UCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUUAGAGGCAUACAUCCGAAACGAAUUUCGCCUUGGCUGCACU ....((((..(((....)))......(((((.((((((((.((.....)))))))))).......((((((......)))))))))))....((((..((.....))..))))))))... ( -30.80) >DroEre_CAF1 5562 120 + 1 UCAAGUAGAACGGAAACCCGAAAUCCAUGCCAAUGGGUUAACUAGUCCAGUGACCUUUGCGUUGCUCUAAAAUGAACUUUAGAGAAAUAAAGCCGAAACAAAUUUCGCCUUGGCUGCACU ...(((....(((....))).(((((((....))))))).)))(((.((((.........(((.(((((((......))))))).))).(((.(((((....))))).))).)))).))) ( -26.80) >DroYak_CAF1 5487 120 + 1 UCAAGUAGAACGGAAACCCGAAAUCCGUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUGCUCUAAAAUGAACUUUAGAGAAAUAAAGCCGAAACAAAUUUCGCCUUGGCUGCACU ....(((..(((((.........)))))(((.((((((((.((.....))))))))))..(((.(((((((......))))))).))).(((.(((((....))))).)))))))))... ( -28.50) >consensus UCAAGCAGAACGGAAACCCGAAAUCCAUGCCAAAGGGUUAACUAGUCAAGUGACCUUUGCGUUACUCUAAAAUGAACUUUAGAGAAAUAAAGCCGAAACGAAUUUCGCCUUGGCUGCACU ....(((...(((....)))........((((((((((((.((.....))))))))))((....(((((((......)))))))..........((((....))))))..)))))))... (-27.08 = -27.04 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:09 2006