
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,139,158 – 19,139,308 |
| Length | 150 |
| Max. P | 0.991543 |

| Location | 19,139,158 – 19,139,278 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19139158 120 + 23771897 ACAUAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGUCAACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGCGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGUU .....((((((.(((.(((((((((((......))))))............(((((((..(......)..)))))))....)))))..)))))))))(((((.....)))))........ ( -28.50) >DroSec_CAF1 2169 120 + 1 ACAAAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGACAACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCU .....((((((.((((((((((.((((......))))..............(((((((..(......)..)))))))...))))))).)))))))))(((((.....)))))........ ( -26.60) >DroSim_CAF1 1707 120 + 1 ACAAAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGACAACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCU .....((((((.((((((((((.((((......))))..............(((((((..(......)..)))))))...))))))).)))))))))(((((.....)))))........ ( -26.60) >DroEre_CAF1 2385 120 + 1 ACAUAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGCCAACGAGUUACAAAUGAGUUUGAUCAAUUUAAUGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCU .....((((((.((((((((((.((((......))))))............((((((((((.....)))).)))))).....))))).)))))))))(((((.....)))))........ ( -25.90) >DroYak_CAF1 2453 120 + 1 ACUUAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGCCAACGAGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAAAGAUCAUUAAUCUCACUGAGUU .......((((..((.....((.((((......))))))...(((......(((((((..(......)..))))))).....(((((((((........))))))))).)))))..)))) ( -25.30) >consensus ACAUAUCAAUUUGGUACCAAGGACUUAAAGACCUAAGACAACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCU .....((((((...((((((((.((((......))))..............(((((((..(......)..)))))))...))))))))...))))))(((((.....)))))........ (-23.36 = -23.96 + 0.60)


| Location | 19,139,198 – 19,139,308 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19139198 110 + 23771897 ACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGCGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGUUGUAAGAUGAAUUAA----------CCAUCAAGUGGUACAG .(((.((((((.(.(((....((((((....((.((.......))))....))))))(((((.....)))))))).).))))))((((......----------.))))..)))...... ( -23.10) >DroSec_CAF1 2209 119 + 1 ACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCUGUAAGAUGAAUUAACCAUCAUUAACCAUCAAGUGGUAC-G ..........((((((((((.(((.((((.((((((......(((((((((........)))))))))......)))))).)))).))).)))))..))))).(((((...)))))..-. ( -29.50) >DroSim_CAF1 1747 109 + 1 ACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCUGUAAGAAGAAUUAA----------CCAUCAAGUGGUAC-G .((..((((..(((.(((((.((..((((.((((((......(((((((((........)))))))))......)))))).))))..)).))))----------))))...))))..)-) ( -26.90) >DroEre_CAF1 2425 109 + 1 ACGAGUUACAAAUGAGUUUGAUCAAUUUAAUGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCUGUAAGAUGAAUUAA----------CCAUCAAGUGGUAC-G ....((..((..(((..(((((((.((((..(((((......(((((((((........)))))))))......)))))..)))).)).)))))----------...)))..))..))-. ( -23.40) >DroYak_CAF1 2493 109 + 1 ACGAGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAAAGAUCAUUAAUCUCACUGAGUUGUAAGAUGAAUUAA----------CCAUCAAGUGGUAC-G .((..((((..(((.(((((.(((.((((.((((((......(((((((((........)))))))))......)))))).)))).))).))))----------))))...))))..)-) ( -25.70) >consensus ACGCGUUACAAAUGAGUUGGAUCAAUUUAAAGCUCAUAAAUCUUGGUGAUCAAUUGAGAGAUCAUUAAUCUCACUGAGCUGUAAGAUGAAUUAA__________CCAUCAAGUGGUAC_G .(((.(((((..(((((((...)))))))..(((((......(((((((((........)))))))))......))))))))))((((.................))))..)))...... (-19.39 = -20.15 + 0.76)

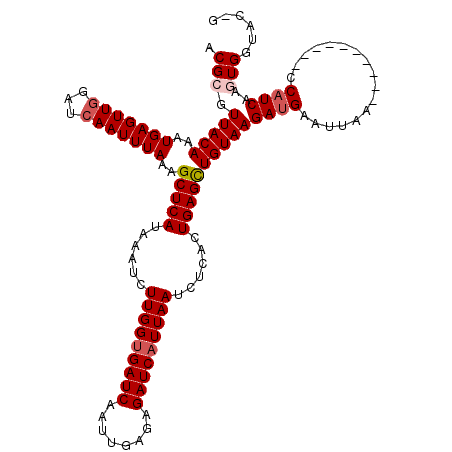
| Location | 19,139,198 – 19,139,308 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19139198 110 - 23771897 CUGUACCACUUGAUGG----------UUAAUUCAUCUUACAACUCAGUGAGAUUAAUGAUCUCUCAAUUGAUCGCCAAGAUUUAUGAGCUUUAAAUUGAUCCAACUCAUUUGUAACGCGU ..((.......(((((----------.....)))))((((((.(.(((..(((((((.....((((...((((.....))))..))))......)))))))..))).).)))))).)).. ( -21.50) >DroSec_CAF1 2209 119 - 1 C-GUACCACUUGAUGGUUAAUGAUGGUUAAUUCAUCUUACAGCUCAGUGAGAUUAAUGAUCUCUCAAUUGAUCACCAAGAUUUAUGAGCUUUAAAUUGAUCCAACUCAUUUGUAACGCGU (-((..((..((((((((((((((((.....)))))(((.((((((...(((((..(((((........)))))....))))).)))))).)))))))).)))..)))..))..)))... ( -28.10) >DroSim_CAF1 1747 109 - 1 C-GUACCACUUGAUGG----------UUAAUUCUUCUUACAGCUCAGUGAGAUUAAUGAUCUCUCAAUUGAUCACCAAGAUUUAUGAGCUUUAAAUUGAUCCAACUCAUUUGUAACGCGU (-((..((..((((((----------((((((.....((.((((((...(((((..(((((........)))))....))))).)))))).)))))))).)))..)))..))..)))... ( -23.00) >DroEre_CAF1 2425 109 - 1 C-GUACCACUUGAUGG----------UUAAUUCAUCUUACAGCUCAGUGAGAUUAAUGAUCUCUCAAUUGAUCACCAAGAUUUAUGAGCAUUAAAUUGAUCAAACUCAUUUGUAACUCGU .-((..((..((((((----------((((((.(((((((......)))))))(((((....((((...((((.....))))..))))))))))))))))))...)))..))..)).... ( -21.40) >DroYak_CAF1 2493 109 - 1 C-GUACCACUUGAUGG----------UUAAUUCAUCUUACAACUCAGUGAGAUUAAUGAUCUUUCAAUUGAUCACCAAGAUUUAUGAGCUUUAAAUUGAUCCAACUCAUUUGUAACUCGU .-......((((.(((----------(((((..(((((((......)))))))...(((....))))))))))).))))....(((((...(((((.((......)))))))...))))) ( -21.40) >consensus C_GUACCACUUGAUGG__________UUAAUUCAUCUUACAGCUCAGUGAGAUUAAUGAUCUCUCAAUUGAUCACCAAGAUUUAUGAGCUUUAAAUUGAUCCAACUCAUUUGUAACGCGU ...........(((((...............)))))((((((.(.(((..(((((((.....((((...((((.....))))..))))......)))))))..))).).))))))..... (-17.54 = -17.34 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:04 2006