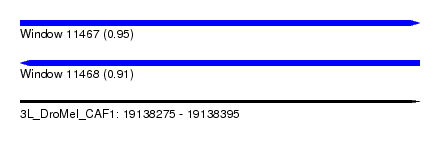
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,138,275 – 19,138,395 |
| Length | 120 |
| Max. P | 0.951504 |

| Location | 19,138,275 – 19,138,395 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
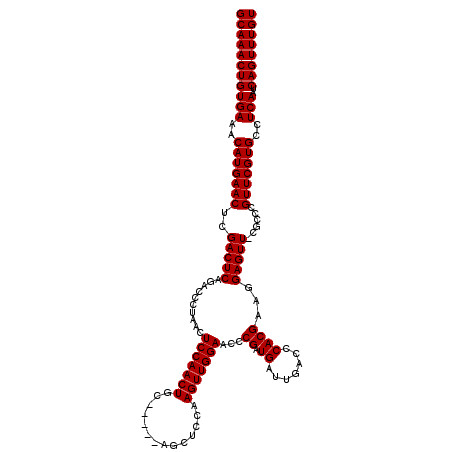
>3L_DroMel_CAF1 19138275 120 + 23771897 GCAAACUGUGAAACAUGAACUCGACUCAGACCCUAACUCCAACUCCCGAGUUGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUUGCGCCCGUUCGUGCCUCAAUCAGUUUGU (((((((((((..(((((((.((((((..........(((((((...(((...)))..)))))))...((.((.......))))...)))))).....)))))))..)))..)))))))) ( -33.80) >DroSec_CAF1 1289 115 + 1 GCAAACUGUGAAACAUGAACUCGACUCAGACCCUAACUCCAACUGC-----AGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUUUCGCCCGUUCGUGCCUCAAUCAGUUUGU (((((((((((..(((((((.(((....((((((...(((((((..-----.......)))))))...((.((.......)))).))).))))))...)))))))..)))..)))))))) ( -32.30) >DroSim_CAF1 834 115 + 1 GCAAACUGUGAAACAUGAACUCGACUCAGACCCUAACUCCAACUGC-----AGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUUUCGCCUGUUCGUGCCUCAAUCAGUUUGU (((((((((((..(((((((.(((....((((((...(((((((..-----.......)))))))...((.((.......)))).))).))))))...)))))))..)))..)))))))) ( -33.00) >DroEre_CAF1 1503 114 + 1 GCAAACUGUGAAACAUGAACUCGACUCGGACGCUAACUCCAACUGC-----AGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUU-CGGCCGUUCGUGCCUCAAUCAGUUUGU (((((((((((..(((((((.(((((.(((.(((............-----)))))).)))))((((((.................)).)))-)....)))))))..)))..)))))))) ( -34.63) >DroYak_CAF1 1550 114 + 1 GCAAACUGUGAAACAUGAACUCGACUCAGACCCUAACUCCAACUGC-----AGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUU-CGGCCGUUCGUGCCUCAAUCAGUUUGU (((((((((((..((((((((((((((..........(((((((..-----.......)))))))...((.((.......))))...))).)-)))..)))))))..)))..)))))))) ( -33.50) >consensus GCAAACUGUGAAACAUGAACUCGACUCAGACCCUAACUCCAACUGC_____AGCUCCAAGUUGGAACCCGAUGAUUGACCCACGAAGGAGUU_CGCCCGUUCGUGCCUCAAUCAGUUUGU (((((((((((..(((((((..(((((..........(((((((..............)))))))...((.((.......))))...)))))......)))))))..)))..)))))))) (-30.32 = -30.32 + 0.00)


| Location | 19,138,275 – 19,138,395 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
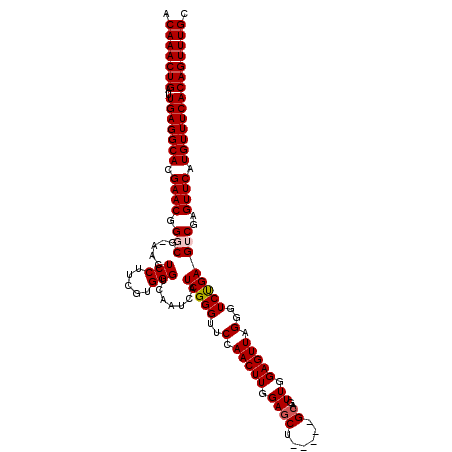
>3L_DroMel_CAF1 19138275 120 - 23771897 ACAAACUGAUUGAGGCACGAACGGGCGCAACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCAACUCGGGAGUUGGAGUUAGGGUCUGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((.....((((((((((((((.....))).)))(((((.....))))).....))))))))((.((((...)))).))..)))).)))))))))))))). ( -43.30) >DroSec_CAF1 1289 115 - 1 ACAAACUGAUUGAGGCACGAACGGGCGAAACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCU-----GCAGUUGGAGUUAGGGUCUGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((.(((((....(((....)))....)).(((((..(.(((((.((.(.-----...))).))))).)..))))))))..)))).)))))))))))))). ( -38.40) >DroSim_CAF1 834 115 - 1 ACAAACUGAUUGAGGCACGAACAGGCGAAACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCU-----GCAGUUGGAGUUAGGGUCUGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((.(((((....(((....)))....)).(((((..(.(((((.((.(.-----...))).))))).)..))))))))..)))).)))))))))))))). ( -38.20) >DroEre_CAF1 1503 114 - 1 ACAAACUGAUUGAGGCACGAACGGCCG-AACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCU-----GCAGUUGGAGUUAGCGUCCGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((((((.-.((......)).)))).....(((((((((.....))))))-----....((((((....).)))))..))))))).)))))))))))))). ( -39.40) >DroYak_CAF1 1550 114 - 1 ACAAACUGAUUGAGGCACGAACGGCCG-AACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCU-----GCAGUUGGAGUUAGGGUCUGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((((((.-((((((..(((((.....))).))((((((.....))))))-----......))))))..))))........)))).)))))))))))))). ( -39.80) >consensus ACAAACUGAUUGAGGCACGAACGGGCG_AACUCCUUCGUGGGUCAAUCAUCGGGUUCCAACUUGGAGCU_____GCAGUUGGAGUUAGGGUCUGAGUCGAGUUCAUGUUUCACAGUUUGC .(((((((..(((((((.((((.(((.....(((.....))).......(((((..(.(((((.((((......))..)).))))).)..))))))))..)))).)))))))))))))). (-35.16 = -35.60 + 0.44)

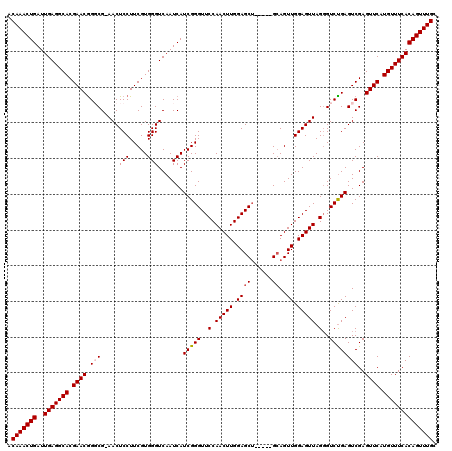
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:00 2006