
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
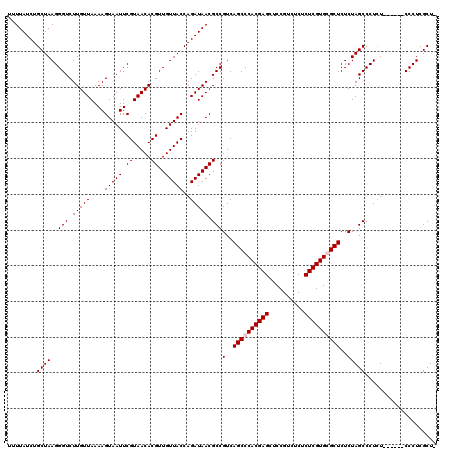
| Location | 19,137,090 – 19,137,243 |
| Length | 153 |
| Max. P | 0.950866 |

| Location | 19,137,090 – 19,137,203 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -26.75 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19137090 113 + 23771897 UUUUAUCUGCUAAGGGUCUUGUUAAAAGUAAUUCGUAACACGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUCU------CCCUCGCU- ........((..((((....((((................(((((((....)))))))..(..(((.((((((..........)))))).)))..))))).....------)))).)).- ( -23.60) >DroSec_CAF1 111 114 + 1 UUUUAUCUGCUAAGGGUCUUGUUAAAAGUAAUUCGUAACACGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUUU------CCCUCGCUA ........((.((((((..(((((.((....))..)))))(((((((....)))))))..(..(((.((((((..........)))))).)))..)..)))))).------.....)).. ( -24.00) >DroEre_CAF1 320 119 + 1 UUUUAUCUGCUAAGGGUCUUGUUAAAAGUAAUUCGUAACACGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUUUCUCUCGUGGGCUCUCUAGCCCUCUCCCUCUCCCUCGCU- ........((((..(((.(((((....(((((.((.....))..)))))..))))).)))(..((((((((((..........))))))))))..)))))...................- ( -30.30) >DroYak_CAF1 321 113 + 1 UUUUAUCUGCUAAGGGUCUUGUUAAAAGUAAUUCGUAACACGUUGUUACCAGAUAACGCCGUCAGCCCACGAGUUCCGUCUCUCUCGUGGGCUCUCUAGCCCUCU------CCCUCGCU- ........((..((((....((((................(((((((....)))))))..(..((((((((((..........))))))))))..))))).....------)))).)).- ( -31.70) >consensus UUUUAUCUGCUAAGGGUCUUGUUAAAAGUAAUUCGUAACACGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUCU______CCCUCGCU_ ........((((..(((.(((((....(((((.((.....))..)))))..))))).)))(..((((((((((..........))))))))))..))))).................... (-26.75 = -27.25 + 0.50)


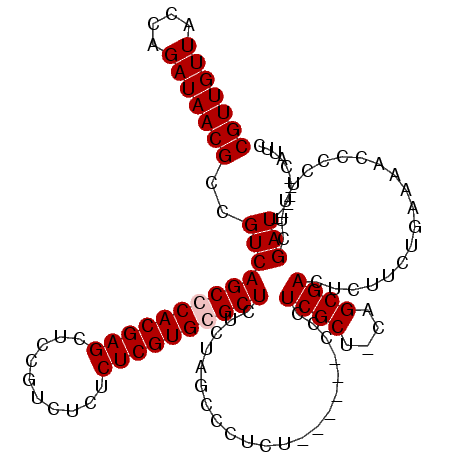
| Location | 19,137,090 – 19,137,203 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -33.73 |
| Energy contribution | -34.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19137090 113 - 23771897 -AGCGAGGG------AGAGGGCUAGAGAGCGCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACGUGUUACGAAUUACUUUUAACAAGACCCUUAGCAGAUAAAA -.(((((((------.....((((((.(((((.(....)...((.((((....))))..))))))).)))))).......(((((.((.....)).)))))...))))).))........ ( -32.90) >DroSec_CAF1 111 114 - 1 UAGCGAGGG------AAAGGGCUAGAGAGCGCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACGUGUUACGAAUUACUUUUAACAAGACCCUUAGCAGAUAAAA ..(((((((------.....((((((.(((((.(....)...((.((((....))))..))))))).)))))).......(((((.((.....)).)))))...))))).))........ ( -32.90) >DroEre_CAF1 320 119 - 1 -AGCGAGGGAGAGGGAGAGGGCUAGAGAGCCCACGAGAGAAACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACGUGUUACGAAUUACUUUUAACAAGACCCUUAGCAGAUAAAA -.(((((((....(((.((.(((.(..((((((((((..........))))))))))..)))).)).))).(((((.....)))))..................))))).))........ ( -39.10) >DroYak_CAF1 321 113 - 1 -AGCGAGGG------AGAGGGCUAGAGAGCCCACGAGAGAGACGGAACUCGUGGGCUGACGGCGUUAUCUGGUAACAACGUGUUACGAAUUACUUUUAACAAGACCCUUAGCAGAUAAAA -.(((((((------.....(((.(..((((((((((..........))))))))))..))))((((...(((((...((.....))..)))))..))))....))))).))........ ( -37.40) >consensus _AGCGAGGG______AGAGGGCUAGAGAGCCCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACGUGUUACGAAUUACUUUUAACAAGACCCUUAGCAGAUAAAA ..(((((((......((((((((.(..((((((((((..........))))))))))..))))........(((((.....)))))......))))).......))))).))........ (-33.73 = -34.05 + 0.31)



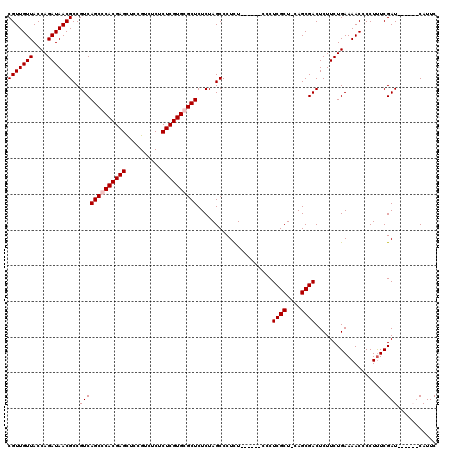
| Location | 19,137,130 – 19,137,243 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -23.60 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19137130 113 + 23771897 CGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUCU------CCCUCGCU-CAGCGACUCUUCUGAAAACCCCUUUCGAUCAUGAUCAUUC (((((((....)))))))..((((((.((((((..........)))))).)))..(((((.....------.....)))-.)).)))......((((.....))))(((....))).... ( -20.00) >DroSec_CAF1 151 114 + 1 CGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUUU------CCCUCGCUACAGCGACUCUUCUGAAAACCCCUUUCGAUCAUGAUCAUUC (((((((....)))))))..((((((.((((((..........)))))).)))((.((((.....------.....)))).)).)))......((((.....))))(((....))).... ( -22.10) >DroEre_CAF1 360 113 + 1 CGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUUUCUCUCGUGGGCUCUCUAGCCCUCUCCCUCUCCCUCGCU-CAGCGACUCUUCUGAAAACCCCUCUCGAU------CAUUC (((((((....)))))))..(((((((((((((..........)))))))))).....................((((.-..))))....................)))------..... ( -25.40) >DroYak_CAF1 361 107 + 1 CGUUGUUACCAGAUAACGCCGUCAGCCCACGAGUUCCGUCUCUCUCGUGGGCUCUCUAGCCCUCU------CCCUCGCU-CAGCGACUCUUCUGAAAACCCCUUUCGAU------CAUUC (((((((....)))))))..(((((((((((((..........))))))))))..(((((.....------.....)))-.)).))).....(((((.....)))))..------..... ( -27.10) >consensus CGUUGUUACCAGAUAACGCCGUCAGCCCACGAGCUCCGUCUCUCUCGUGCGCUCUCUAGCCCUCU______CCCUCGCU_CAGCGACUCUUCUGAAAACCCCUUUCGAU______CAUUC (((((((....)))))))..(((((((((((((..........)))))))))).....................((((....))))....................)))........... (-23.60 = -24.10 + 0.50)

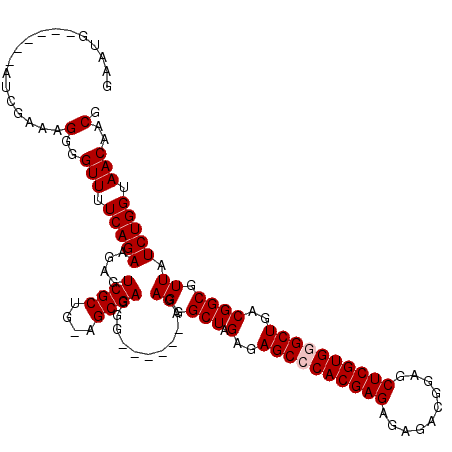
| Location | 19,137,130 – 19,137,243 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -31.83 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19137130 113 - 23771897 GAAUGAUCAUGAUCGAAAGGGGUUUUCAGAAGAGUCGCUG-AGCGAGGG------AGAGGGCUAGAGAGCGCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACG ...((((....))))...(..(((.(((((.....(((((-(((.....------.....)))....(((.((((((..........)))))).)))..)))))...))))).)))..). ( -32.10) >DroSec_CAF1 151 114 - 1 GAAUGAUCAUGAUCGAAAGGGGUUUUCAGAAGAGUCGCUGUAGCGAGGG------AAAGGGCUAGAGAGCGCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACG ...((((....))))...(..(((.(((((.((((((((.((((.....------.....)))).))(((.((((((..........)))))).)))..)))).)).))))).)))..). ( -34.20) >DroEre_CAF1 360 113 - 1 GAAUG------AUCGAGAGGGGUUUUCAGAAGAGUCGCUG-AGCGAGGGAGAGGGAGAGGGCUAGAGAGCCCACGAGAGAAACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACG .....------.......(..(((.(((((.....(((((-(((................)))....((((((((((..........))))))))))..)))))...))))).)))..). ( -35.39) >DroYak_CAF1 361 107 - 1 GAAUG------AUCGAAAGGGGUUUUCAGAAGAGUCGCUG-AGCGAGGG------AGAGGGCUAGAGAGCCCACGAGAGAGACGGAACUCGUGGGCUGACGGCGUUAUCUGGUAACAACG .....------.......(..(((.(((((.....(((((-(((.....------.....)))....((((((((((..........))))))))))..)))))...))))).)))..). ( -35.70) >consensus GAAUG______AUCGAAAGGGGUUUUCAGAAGAGUCGCUG_AGCGAGGG______AGAGGGCUAGAGAGCCCACGAGAGAGACGGAGCUCGUGGGCUGACGGCGUUAUCUGGUAACAACG ..................(..(((.(((((....((((....))))...........((.(((.(..((((((((((..........))))))))))..)))).)).))))).)))..). (-31.83 = -32.33 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:59 2006