
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,135,198 – 19,135,299 |
| Length | 101 |
| Max. P | 0.567650 |

| Location | 19,135,198 – 19,135,299 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19135198 101 - 23771897 UUAUAUGUGUGUCUGUGUAUCCGUCAGAUACUUGCU------GUAUCCGUGUGU-GCCUGCAUCUCCAGGUGUUC-------GUGCGCCAGUG----UGAGUGUUUGAUGUAAGCAGAC ..........((((((.....(((((((((((..(.------....(.(.(((.-.(..(((((....)))))..-------)..)))).).)----..)))))))))))...)))))) ( -35.70) >DroSec_CAF1 22643 101 - 1 UUAUAUGUACAUCUGUGUAUCCGUCAGAUACUUGCU------GUAUCCGUGUGU-GCCUGCAUCUCCAGGUGUUC-------GUGAGCCAGUG----UGAGUGUUUGAUGUAAGCAGAC ...........(((((.....(((((((((((((((------(..((((...(.-.((((......))))..).)-------).))..)))).----.))))))))))))...))))). ( -34.50) >DroSim_CAF1 23570 101 - 1 UUAUAUGUACAUCUGUGUAUCCGUCAGAUACUUGCU------GUAUCCGUGUGU-GCCUGCAUCUCCAGGUGUUC-------GUGCGCCAGUG----UGAGUGUUUGAUGUAAGCAGAC ...........(((((.....(((((((((((..(.------....(.(.(((.-.(..(((((....)))))..-------)..)))).).)----..)))))))))))...))))). ( -33.80) >DroEre_CAF1 16162 104 - 1 UUUUAUGUACAUCUGUGUAUCCGUCAGAUACUUGCUGU---GAUAUCCGUGUGU-GCCUGCAUCUCCAGGUGUUC-------GUGCACCAGUG----UGAGUGUUUGAUGUAAGCAGAC ...........(((((.....(((((((((((..(..(---(........(((.-.(..(((((....)))))..-------)..)))))..)----..)))))))))))...))))). ( -33.60) >DroYak_CAF1 20799 104 - 1 UUAUAAGUACAUCUGUGUAUCCGUCAGAUACUUGCGGC---UGUAUCCGUGUGU-GCCUGCAUCUCCAGGUGUUC-------GUGCAACAGUG----UGAGUGUUUGAUGUAAGCAGAC ...........(((((.....(((((((((((..((.(---(((..(((...(.-.((((......))))..).)-------).)..))))))----..)))))))))))...))))). ( -36.50) >DroAna_CAF1 20974 110 - 1 CUGUGU---------UGUAUCCACCAGAUACUCCCAUCCAGAGUAUCUGUGUGCCGCUUGCAUCUCCAGGUGCUCCCCCUGAGUGUGUGUGUGUGUCAGAGUGUUUGAUGUAAGCAGAC ((((.(---------((((((((((((((((((.......))))))))).))).(((((((((..(((.(..(((.....)))..).)).).))))..)))))...))))))))))).. ( -34.70) >consensus UUAUAUGUACAUCUGUGUAUCCGUCAGAUACUUGCU______GUAUCCGUGUGU_GCCUGCAUCUCCAGGUGUUC_______GUGCGCCAGUG____UGAGUGUUUGAUGUAAGCAGAC ...........(((((.....((((((((((((...............((((.......)))).....(((((...........))))).........))))))))))))...))))). (-20.51 = -20.98 + 0.48)

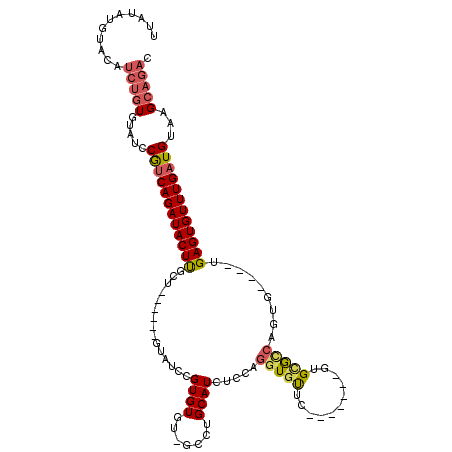

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:52 2006