
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,130,199 – 19,130,358 |
| Length | 159 |
| Max. P | 0.998813 |

| Location | 19,130,199 – 19,130,318 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -26.04 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
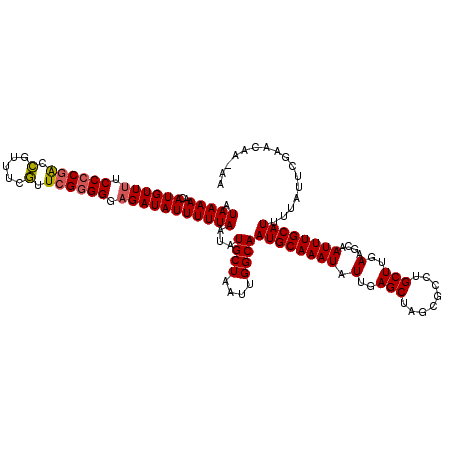
>3L_DroMel_CAF1 19130199 119 + 23771897 UU-UUGUUCGAAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCGAACGAAACGGUCGGGGAGAACAUGUUUUUAU ..-.((((.......(((((((((.(((...(((....)))))).....))))))))).......))))...(((((((((.((((((((.((...)).)))))).))..))))))))). ( -31.44) >DroSec_CAF1 17622 119 + 1 UU-UUGUUCGAAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUGCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAU ..-.((((.......(((((((((.(((...(((....)))))).....))))))))).......))))...(((((((((.(.((((((.((...)).))))))...).))))))))). ( -30.34) >DroSim_CAF1 18495 119 + 1 UU-UUGUUCGAAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUGCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAU ..-.((((.......(((((((((.(((...(((....)))))).....))))))))).......))))...(((((((((.(.((((((.((...)).))))))...).))))))))). ( -30.34) >DroEre_CAF1 11145 119 + 1 UU-UUGUUCGUAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCCAAUGAAAUAGUCGGGGCAAACAUGUUUUUAU ..-.((((.((....(((((((((.(((...(((....)))))).....)))))))))....)).))))...(((((((((....((((.((......)).)))).....))))))))). ( -25.20) >DroYak_CAF1 16064 119 + 1 UUUUUGUUCGAAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCC-CGAAUGAAAUAGCCGGGGAAAACAUGUUUUUAU ....((((.......(((((((((.(((...(((....)))))).....))))))))).......))))...(((((((((.((((-((...........))))))....))))))))). ( -27.84) >consensus UU_UUGUUCGAAUAAAAUGCAAAUUGCUUCAAGCAGGCGCUAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAU ....((((.......(((((((((.(((...(((....)))))).....))))))))).......))))...(((((((((...((((((.((...)).)))))).....))))))))). (-26.04 = -25.92 + -0.12)


| Location | 19,130,199 – 19,130,318 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.72 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19130199 119 - 23771897 AUAAAAACAUGUUCUCCCCGACCGUUUCGUUCGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUUCGAACAA-AA .........(((((.((((((.((...)).))))))(((............((((....))))((((((((.(..(((........)))..)....))))))))....)))))))).-.. ( -32.20) >DroSec_CAF1 17622 119 - 1 AUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGCAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUUCGAACAA-AA .(((((..((((((.((((((.((...)).)))))).)))))))))))...((((....))))((((((((.(..(((........)))..)....)))))))).............-.. ( -30.10) >DroSim_CAF1 18495 119 - 1 AUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGCAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUUCGAACAA-AA .(((((..((((((.((((((.((...)).)))))).)))))))))))...((((....))))((((((((.(..(((........)))..)....)))))))).............-.. ( -30.10) >DroEre_CAF1 11145 119 - 1 AUAAAAACAUGUUUGCCCCGACUAUUUCAUUGGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUACGAACAA-AA .........((((((((((((........))))))................((((....))))((((((((.(..(((........)))..)....))))))))......)))))).-.. ( -27.70) >DroYak_CAF1 16064 119 - 1 AUAAAAACAUGUUUUCCCCGGCUAUUUCAUUCG-GGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUUCGAACAAAAA .((((((.....(((((((((.........)))-))))))..))))))...((((....))))((((((((.(..(((........)))..)....))))))))................ ( -27.70) >consensus AUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUAGCGCCUGCUUGAAGCAAUUUGCAUUUUAUUCGAACAA_AA .(((((..((((((.((((((.(.....).)))))).)))))))))))...((((....))))((((((((.(..(((........)))..)....))))))))................ (-26.76 = -26.72 + -0.04)


| Location | 19,130,238 – 19,130,358 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19130238 120 + 23771897 UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCGAACGAAACGGUCGGGGAGAACAUGUUUUUAUCGCUUUUUCAUGUGGGCUCUAUUUGCCCAAACUCGAAAUA .(((.............(((......)))...(((((((((.((((((((.((...)).)))))).))..)))))))))..))).((((...(((((.......))))).....)))).. ( -30.60) >DroSec_CAF1 17661 120 + 1 UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUGCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAUCGCUUUUUCAUGUGGGCUCUGUUUGCCCAAACUCGAAAUA .(((.............(((......)))...(((((((((.(.((((((.((...)).))))))...).)))))))))..))).((((...(((((.......))))).....)))).. ( -29.50) >DroSim_CAF1 18534 120 + 1 UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUGCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAUCGCUUUUUCAUGUGGGCUCUGUUUGCCCAAACUCGAAAUA .(((.............(((......)))...(((((((((.(.((((((.((...)).))))))...).)))))))))..))).((((...(((((.......))))).....)))).. ( -29.50) >DroEre_CAF1 11184 120 + 1 UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCCAAUGAAAUAGUCGGGGCAAACAUGUUUUUAUCGUUUUUUCAUGUGGGCUCUGUUUGCCCAAGCUCGAAAUA .((((........(((((((......)))...(((((((((....((((.((......)).)))).....)))))))))..........))))((((.......)))).))))....... ( -24.00) >DroYak_CAF1 16104 119 + 1 UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCC-CGAAUGAAAUAGCCGGGGAAAACAUGUUUUUAUCGCUUUUUCAUGUGGGCUCUGUUUGCCCAAACUCGAAAUA .(((.............(((......)))...(((((((((.((((-((...........))))))....)))))))))..))).((((...(((((.......))))).....)))).. ( -27.00) >consensus UAGCUCAAUAUUUGCAUUGCCAAUUAGCAUAUUAAAAAUAUCUCCCCCGAACGAAACGGUCGGGGAAAACAUGUUUUUAUCGCUUUUUCAUGUGGGCUCUGUUUGCCCAAACUCGAAAUA .(((.............(((......)))...(((((((((...((((((.((...)).)))))).....)))))))))..))).((((...(((((.......))))).....)))).. (-24.88 = -24.60 + -0.28)



| Location | 19,130,238 – 19,130,358 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19130238 120 - 23771897 UAUUUCGAGUUUGGGCAAAUAGAGCCCACAUGAAAAAGCGAUAAAAACAUGUUCUCCCCGACCGUUUCGUUCGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA ..(((((.((..((((.......)))))).))))).(((..(((((..(((((..((((((.((...)).))))))..))))))))))...((((....)))).............))). ( -33.00) >DroSec_CAF1 17661 120 - 1 UAUUUCGAGUUUGGGCAAACAGAGCCCACAUGAAAAAGCGAUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGCAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA ..(((((.((..((((.......)))))).))))).(((..(((((..((((((.((((((.((...)).)))))).)))))))))))...((((....)))).............))). ( -31.40) >DroSim_CAF1 18534 120 - 1 UAUUUCGAGUUUGGGCAAACAGAGCCCACAUGAAAAAGCGAUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGCAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA ..(((((.((..((((.......)))))).))))).(((..(((((..((((((.((((((.((...)).)))))).)))))))))))...((((....)))).............))). ( -31.40) >DroEre_CAF1 11184 120 - 1 UAUUUCGAGCUUGGGCAAACAGAGCCCACAUGAAAAAACGAUAAAAACAUGUUUGCCCCGACUAUUUCAUUGGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA .......(((((((((.......)))))..........(((((.(((.((((((.((((.............)))).)))))).)))....((((....)))).......))))))))). ( -29.02) >DroYak_CAF1 16104 119 - 1 UAUUUCGAGUUUGGGCAAACAGAGCCCACAUGAAAAAGCGAUAAAAACAUGUUUUCCCCGGCUAUUUCAUUCG-GGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA ..(((((.((..((((.......)))))).))))).(((..((((((.....(((((((((.........)))-))))))..))))))...((((....)))).............))). ( -29.00) >consensus UAUUUCGAGUUUGGGCAAACAGAGCCCACAUGAAAAAGCGAUAAAAACAUGUUUUCCCCGACCGUUUCGUUCGGGGGAGAUAUUUUUAAUAUGCUAAUUGGCAAUGCAAAUAUUGAGCUA .......(((((((((.......))))..........(((.(((((..((((((.((((((.(.....).)))))).)))))))))))...((((....)))).))).......))))). (-27.34 = -27.34 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:48 2006