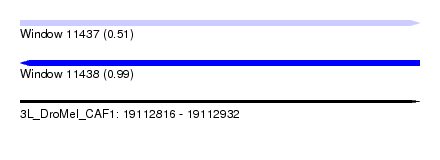
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,112,816 – 19,112,932 |
| Length | 116 |
| Max. P | 0.994747 |

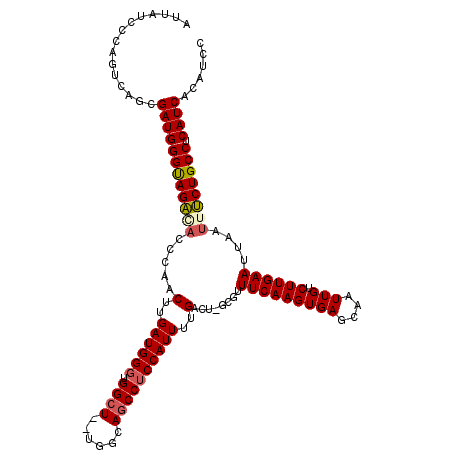
| Location | 19,112,816 – 19,112,932 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19112816 116 + 23771897 GGAUGUGAUGAGGCAGAAAUUAAUUCAAGACAAUUGCUCACUUGAAAUGC-AGUCAAAAUGGAGGCUGCCA--AGCCACCCAUCAAGUUGGGUCUCUACCCAUCGCUGACUGGGAUAAU ...........(((.........((((((...........))))))..((-((((........))))))..--.))).(((((((.(.(((((....))))).)..))).))))..... ( -32.70) >DroSec_CAF1 346 116 + 1 GGAUGUGAUGAGGCAGAAAUUAAUUCAAGACAAUUGCUCACUUGAAACGC-AGUCAAAAUGGAGGCUGCCA--AGCCACCCAUCAAGUUGGGUGUCUACCCAUCGCUGACUGGGAUAAU ....((((((.((.(((......((((((...........))))))..((-((((........))))))..--...((((((......))))))))).))))))))............. ( -34.10) >DroSim_CAF1 1258 116 + 1 GGAUGUGAUGAGGCAGAAAUUAAUUCAAGACAAUUGCUCACUUGAAACGC-AGUCAAAAUGGAGGCUGCCA--AGCCACCCAUCAAGUUGGGUGUCUACCCAUCGCUGACUGGGAUAAU ....((((((.((.(((......((((((...........))))))..((-((((........))))))..--...((((((......))))))))).))))))))............. ( -34.10) >DroYak_CAF1 1384 116 + 1 GGAUGUGAUGAGGCAGAAAUUAAUUCAAGACAAUUGCUCACUUGAAACGC-AGUCAAAAUGGAGGCUGCAA--AGCCACCCAUCAAGUUGGCUCUCUACCCAUCAUUGACUGGGGCAAU ...(((.....(((.........((((((...........))))))..((-((((........))))))..--.))).((((((((..(((........)))...)))).))))))).. ( -31.90) >DroAna_CAF1 383 111 + 1 GGAUGUGAUGGGGCAGGAAUUAAUUCAAGACAAUUGCUCACUUGAAACGAAAGUCAAAAUGGUGGUUGCUGAGAGCCACCCAUCAAGUG----UCCUGCCCAUCUCCCACU----UAAU (((...((((((.(((((.......(((.....)))..(((((((.((....))......(((((((......)))))))..)))))))----))))))))))))))....----.... ( -39.70) >consensus GGAUGUGAUGAGGCAGAAAUUAAUUCAAGACAAUUGCUCACUUGAAACGC_AGUCAAAAUGGAGGCUGCCA__AGCCACCCAUCAAGUUGGGUCUCUACCCAUCGCUGACUGGGAUAAU ....((((((.((.(((........(((.....)))((.((((((.((....))......((.((((......)))).))..)))))).))...))).))))))))............. (-23.40 = -23.08 + -0.32)

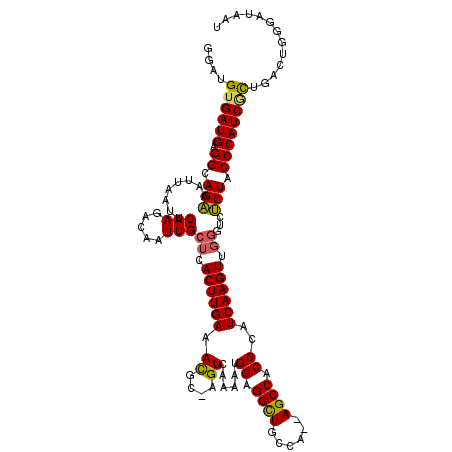
| Location | 19,112,816 – 19,112,932 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19112816 116 - 23771897 AUUAUCCCAGUCAGCGAUGGGUAGAGACCCAACUUGAUGGGUGGCU--UGGCAGCCUCCAUUUUGACU-GCAUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUUCUGCCUCAUCACAUCC .............(.((((((((((((.....(..((((((.((((--....))))))))))..)...-....(((((((((....))).))))))....)))))))).)))).).... ( -33.90) >DroSec_CAF1 346 116 - 1 AUUAUCCCAGUCAGCGAUGGGUAGACACCCAACUUGAUGGGUGGCU--UGGCAGCCUCCAUUUUGACU-GCGUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUUCUGCCUCAUCACAUCC .............(.((((((((((((((((......))))))...--..((((.(........).))-))..(((((((((....))).))))))......)))))).)))).).... ( -34.30) >DroSim_CAF1 1258 116 - 1 AUUAUCCCAGUCAGCGAUGGGUAGACACCCAACUUGAUGGGUGGCU--UGGCAGCCUCCAUUUUGACU-GCGUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUUCUGCCUCAUCACAUCC .............(.((((((((((((((((......))))))...--..((((.(........).))-))..(((((((((....))).))))))......)))))).)))).).... ( -34.30) >DroYak_CAF1 1384 116 - 1 AUUGCCCCAGUCAAUGAUGGGUAGAGAGCCAACUUGAUGGGUGGCU--UUGCAGCCUCCAUUUUGACU-GCGUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUUCUGCCUCAUCACAUCC ..............(((((((((((((((...(..((((((.((((--....))))))))))..)...-))..(((((((((....))).))))))....)))))))).)))))..... ( -36.00) >DroAna_CAF1 383 111 - 1 AUUA----AGUGGGAGAUGGGCAGGA----CACUUGAUGGGUGGCUCUCAGCAACCACCAUUUUGACUUUCGUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUCCUGCCCCAUCACAUCC ....----.((((.....((((((((----(((((((..(((((..........))))).....(((....))))))))))..(((.....))).......))))))))..)))).... ( -34.30) >consensus AUUAUCCCAGUCAGCGAUGGGUAGACACCCAACUUGAUGGGUGGCU__UGGCAGCCUCCAUUUUGACU_GCGUUUCAAGUGAGCAAUUGUCUUGAAUUAAUUUCUGCCUCAUCACAUCC ...............((((((((((((.....(..((((((.((((......))))))))))..)........(((((((((....))).))))))....)))))))).))))...... (-31.20 = -31.64 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:31 2006