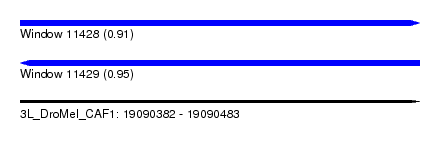
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,090,382 – 19,090,483 |
| Length | 101 |
| Max. P | 0.951513 |

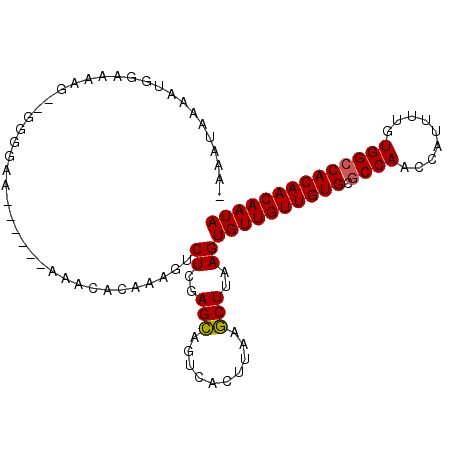
| Location | 19,090,382 – 19,090,483 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -19.03 |
| Energy contribution | -18.75 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19090382 101 + 23771897 -AAAUAAAAGGGCAAGG--GGGAAAAAACCAAAACUCAAAGUCUCGAGCAGUCACUUAAGCUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA -........(((....(--(........))....))).....((..(((..........)))..))((((((((((.((((.........)))))))))))))) ( -25.00) >DroSim_CAF1 7854 101 + 1 -AAAUAAAAUGGAAAGG--AGGAAACAACAAAAACUCAAAGUCUCGAGCAGUCACUUAAGCUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA -..(((((((((...((--.....(((((((...(((........)))....((((((....)))))))))))))...))..)))))))))............. ( -22.90) >DroEre_CAF1 8529 95 + 1 -AAAUAAAAUGGAAAAG--GGGGAA------GAACACAAAGUCUCGAGCAGUCACUUAAGCUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA -................--......------.(((((.(((.((..((......))..)))))..)))))((((((.((((.........)))))))))).... ( -21.20) >DroWil_CAF1 15065 95 + 1 UCAUUAUGAUGUUGGUU--GUCG------U-CUCACAAAAGUCUCGAGUAGUCACUUAAACUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA .......((((......--..))------)-).(((..((((.(..(((....)))..)))))..)))((((((((.((((.........)))))))))))).. ( -22.10) >DroYak_CAF1 7756 97 + 1 -AAAUAAAAUGGGAAAAGAGGGGGA------AAAGAGAAAGUCUCGAGCAGUCACUUAAGCUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA -........((((..(((.((..(.------...(((.....)))...)..)).)))...))))..((((((((((.((((.........)))))))))))))) ( -23.40) >DroAna_CAF1 8908 96 + 1 -ACACAAA-CACACACACACAG-----ACC-AAGUCAAAAGUCUCGAGUAGUCACUUAAACUUAAGUGUUGUUGUGCACCAACCAUUUUGUGGCCACAACAAUA -.....((-(((.........(-----((.-..)))..((((.(..(((....)))..)))))..)))))((((((..(((.........))).)))))).... ( -16.60) >consensus _AAAUAAAAUGGAAAAG__GGGGAA______AAACACAAAGUCUCGAGCAGUCACUUAAGCUUAAGUGUUGUUGUGCGCCAACCAUUUUGUGGCCACAACAAUA ..........................................((..(((..........)))..))((((((((((.((((.........)))))))))))))) (-19.03 = -18.75 + -0.28)

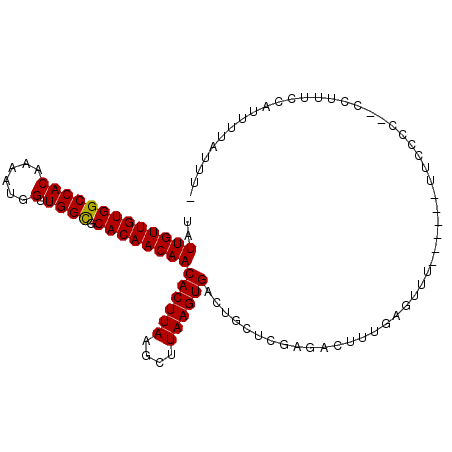
| Location | 19,090,382 – 19,090,483 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -20.09 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19090382 101 - 23771897 UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGCUUAAGUGACUGCUCGAGACUUUGAGUUUUGGUUUUUUCCC--CCUUGCCCUUUUAUUU- ..(((((((((((((......).)))).))))))))((((((....))))))...((.((((((.....))))))))........--................- ( -25.10) >DroSim_CAF1 7854 101 - 1 UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGCUUAAGUGACUGCUCGAGACUUUGAGUUUUUGUUGUUUCCU--CCUUUCCAUUUUAUUU- ..(((((((((((((......).)))).))))))))((((((....))))))...((((((....))))))..............--................- ( -24.10) >DroEre_CAF1 8529 95 - 1 UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGCUUAAGUGACUGCUCGAGACUUUGUGUUC------UUCCCC--CUUUUCCAUUUUAUUU- ...((((((((((((......).)))).)))))))((((..(((((..(((....)))..)).))).))))..------......--................- ( -24.10) >DroWil_CAF1 15065 95 - 1 UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGUUUAAGUGACUACUCGAGACUUUUGUGAG-A------CGAC--AACCAACAUCAUAAUGA ..(((((((((((((......).)))).))))))))(((..((((((.(((....)))..))))))..)))..-.------....--................. ( -24.00) >DroYak_CAF1 7756 97 - 1 UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGCUUAAGUGACUGCUCGAGACUUUCUCUUU------UCCCCCUCUUUUCCCAUUUUAUUU- ..(((((((((((((......).)))).))))))))((((((....)))))).......(((.....)))...------........................- ( -22.90) >DroAna_CAF1 8908 96 - 1 UAUUGUUGUGGCCACAAAAUGGUUGGUGCACAACAACACUUAAGUUUAAGUGACUACUCGAGACUUUUGACUU-GGU-----CUGUGUGUGUGUG-UUUGUGU- ..(((((((((((((......).)))).))))))))((((((....))))))(((((.(((((((........-)))-----)).)).))).)).-.......- ( -25.20) >consensus UAUUGUUGUGGCCACAAAAUGGUUGGCGCACAACAACACUUAAGCUUAAGUGACUGCUCGAGACUUUGAGUUU______UUCCCC__CCUUUCCAUUUUAUUU_ ..(((((((((((((......).)))).))))))))((((((....)))))).................................................... (-20.09 = -19.95 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:22 2006