
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,090,241 – 19,090,364 |
| Length | 123 |
| Max. P | 0.879531 |

| Location | 19,090,241 – 19,090,332 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -27.14 |
| Energy contribution | -28.67 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19090241 91 + 23771897 UUGUCUACGUGGCAAAACUUCCGCUGCGGUU-UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGUGGCUGCGGCAACCGGAAGCAGCAAACAC .......((..((.........))..))(((-...((((((.((((.((..(((((((....)))))))..)).))))..)))))).))).. ( -41.30) >DroPse_CAF1 11378 74 + 1 UUGUCUACGUGGCAGGACUUCCGCUGCUGUUGUCGGCUGCUGCG------------------GUGGCUGCGGCAACCGGAAACGCCAAACAC .........((((.((....))......(((..(((.(((((((------------------(...)))))))).)))..)))))))..... ( -27.90) >DroSim_CAF1 7713 91 + 1 UUGUCUACGUGGCAAAACUUCCGCUGCGGUU-UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGUGGCUGCGGCAACCGGAAGCAGCAAACAC .......((..((.........))..))(((-...((((((.((((.((..(((((((....)))))))..)).))))..)))))).))).. ( -41.30) >DroEre_CAF1 8390 91 + 1 UUGUCUACGUGGCAAAACUUCCGCUGCGGUU-UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGUGGCUGCGGCAGCCGGAAGCACCAAACAC (((((.....)))))..((((((((((.((.-..(((..(((.((((((....)))))).)))..))))).)))).)))))).......... ( -41.70) >DroYak_CAF1 7617 91 + 1 UUGUCUACGUGGCAAAACUUCCGCUGCGGUU-UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGUGGCUGCGGCAGCCGGAAGUACCAAACAC (((((.....))))).(((((((((((.((.-..(((..(((.((((((....)))))).)))..))))).)))).)))))))......... ( -43.50) >DroPer_CAF1 6775 74 + 1 UUGUCUACGUGGCAGGACUUCCGCUGCUGUUGUCGGCUGCUGCG------------------GUGGCUGCGGCAACCGGAAACGCCAAACAC .........((((.((....))......(((..(((.(((((((------------------(...)))))))).)))..)))))))..... ( -27.90) >consensus UUGUCUACGUGGCAAAACUUCCGCUGCGGUU_UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGUGGCUGCGGCAACCGGAAGCACCAAACAC (((((.....)))))..((((((.(((.((....(((..(((.((((((....)))))).)))..))))).)))..)))))).......... (-27.14 = -28.67 + 1.53)


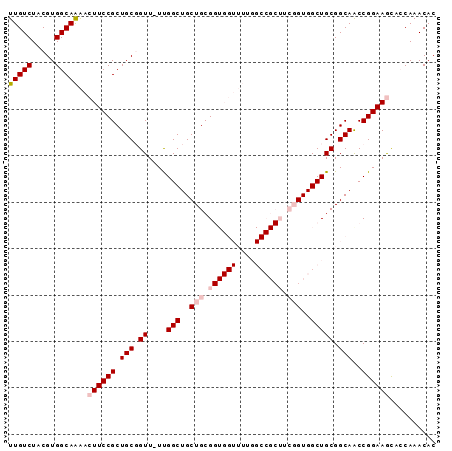
| Location | 19,090,272 – 19,090,364 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -30.53 |
| Energy contribution | -30.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
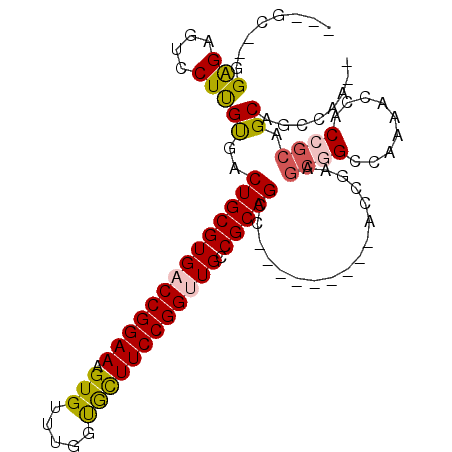
>3L_DroMel_CAF1 19090272 92 + 23771897 --UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGU---------GGCUGCGGCAACCGGAAGCAGCAAACACUUUCCGGUCACGCAGUCACAAGGACUCUCC--GC--- --...((((((.((((.((..(((((((....))---------)))))..)).))))..))))))..........(((....(.((((.....))))).))--).--- ( -38.30) >DroSim_CAF1 7744 92 + 1 --UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGU---------GGCUGCGGCAACCGGAAGCAGCAAACACUUUCCGGUCACGCAGUCACAAGGACUCUCC--GC--- --...((((((.((((.((..(((((((....))---------)))))..)).))))..))))))..........(((....(.((((.....))))).))--).--- ( -38.30) >DroEre_CAF1 8421 92 + 1 --UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGU---------GGCUGCGGCAGCCGGAAGCACCAAACACUUUCCGGUCACGCAGUCACAAGGACUCUCC--GC--- --.((((((((((.((......((((((....))---------)))))).)))((((((((..........))))))))...)))))))...((.....))--..--- ( -37.80) >DroYak_CAF1 7648 92 + 1 --UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGU---------GGCUGCGGCAGCCGGAAGUACCAAACACUUUCCGGUCACGCAGUCGCAAGGACUCUCC--GC--- --...((.((((((((.((...((((((....))---------)))))).)).(((((((((.......)).)))))))..)))))).))..((.....))--..--- ( -40.40) >DroAna_CAF1 8782 103 + 1 GGUUGGCUGCUGCGGUGGAUUUGCCGGCUUUGGUGGCUUUGGCGGCUGCGGCAACCGGAAGUGCCAAACACUUUCCGGUCACGCAGUCACGAGGUUGCCCU--CC--- (((((((..((.....))....)))))))..((..((((((..(((((((...((((((((((.....))).)))))))..))))))).))))))..))..--..--- ( -48.80) >DroPer_CAF1 6806 80 + 1 -GUCGGCUGCUGCG------------------GU---------GGCUGCGGCAACCGGAAACGCCAAACACUUUCCGGUCACGCAGUCACGAGGUUUCCCUCAGCUGU -..(((((......------------------((---------(((((((...((((((((..........))))))))..)))))))))((((....))))))))). ( -38.00) >consensus __UUGGCUGCUGCGGUGGUUUUGGCCGCUUCGGU_________GGCUGCGGCAACCGGAAGCACCAAACACUUUCCGGUCACGCAGUCACAAGGACUCUCC__GC___ ..(((...((((.(((((......))))).)))).........(((((((...((((((((..........))))))))..))))))).)))................ (-30.53 = -30.53 + 0.00)



| Location | 19,090,272 – 19,090,364 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
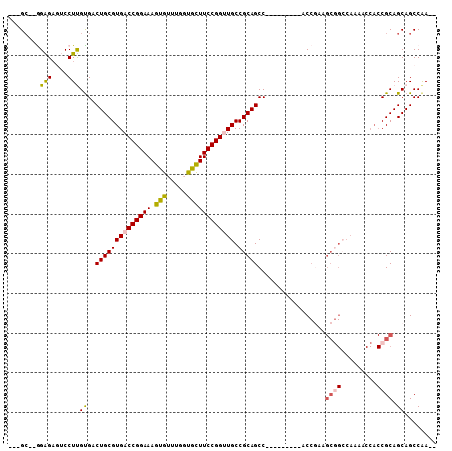
>3L_DroMel_CAF1 19090272 92 - 23771897 ---GC--GGAGAGUCCUUGUGACUGCGUGACCGGAAAGUGUUUGCUGCUUCCGGUUGCCGCAGCC---------ACCGAAGCGGCCAAAACCACCGCAGCAGCCAA-- ---((--((.(..((...(((.(((((..((((((((((....)))..)))))))..).)))).)---------)).))..)((......)).)))).........-- ( -35.30) >DroSim_CAF1 7744 92 - 1 ---GC--GGAGAGUCCUUGUGACUGCGUGACCGGAAAGUGUUUGCUGCUUCCGGUUGCCGCAGCC---------ACCGAAGCGGCCAAAACCACCGCAGCAGCCAA-- ---((--((.(..((...(((.(((((..((((((((((....)))..)))))))..).)))).)---------)).))..)((......)).)))).........-- ( -35.30) >DroEre_CAF1 8421 92 - 1 ---GC--GGAGAGUCCUUGUGACUGCGUGACCGGAAAGUGUUUGGUGCUUCCGGCUGCCGCAGCC---------ACCGAAGCGGCCAAAACCACCGCAGCAGCCAA-- ---((--((.(..((...(((.((((((..((((((.((.......))))))))..).))))).)---------)).))..)((......)).)))).........-- ( -31.10) >DroYak_CAF1 7648 92 - 1 ---GC--GGAGAGUCCUUGCGACUGCGUGACCGGAAAGUGUUUGGUACUUCCGGCUGCCGCAGCC---------ACCGAAGCGGCCAAAACCACCGCAGCAGCCAA-- ---..--((.....))..((..(((((((.((((((.(((.....)))))))))..(((((....---------......)))))......)).)))))..))...-- ( -31.00) >DroAna_CAF1 8782 103 - 1 ---GG--AGGGCAACCUCGUGACUGCGUGACCGGAAAGUGUUUGGCACUUCCGGUUGCCGCAGCCGCCAAAGCCACCAAAGCCGGCAAAUCCACCGCAGCAGCCAACC ---.(--(((....))))(((.(((((..(((((((.(((.....))))))))))..).)))).)))....(((.........))).........((....))..... ( -41.30) >DroPer_CAF1 6806 80 - 1 ACAGCUGAGGGAAACCUCGUGACUGCGUGACCGGAAAGUGUUUGGCGUUUCCGGUUGCCGCAGCC---------AC------------------CGCAGCAGCCGAC- ...(((((((....))))(((.(((((..((((((((.((.....))))))))))..).)))).)---------))------------------...))).......- ( -38.70) >consensus ___GC__GGAGAGUCCUUGUGACUGCGUGACCGGAAAGUGUUUGGUGCUUCCGGUUGCCGCAGCC_________ACCGAAGCGGCCAAAACCACCGCAGCAGCCAA__ ........(((....)))((..((((((((((((((.(((.....)))))))))))).))))).................((((.........)))).))........ (-23.56 = -23.43 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:20 2006