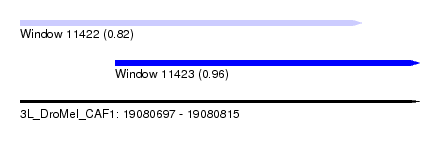
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,080,697 – 19,080,815 |
| Length | 118 |
| Max. P | 0.957114 |

| Location | 19,080,697 – 19,080,798 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.54 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -8.44 |
| Energy contribution | -8.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19080697 101 + 23771897 UCUA-------CCCACCA-AUGGGAUU----CCACCAUAGAGUCUAGUUGGCCUAGAUGAGGAAAUUCCACUAUCACAAAGGCCGCAACAACUUUA------ACACCAGAAGAAAAUCC (((.-------((((...-.))))...----......((((((.....((((((.((((.((.....))..))))....)))))).....))))))------.....)))......... ( -20.50) >DroSec_CAF1 5685 100 + 1 UCUA-------CCAACCA-AUGGGAUU----CCACCAUAGAGUCUAGUUGGCCUAGAUGAGGAAAU-CCUCCAUUACAAAGGCCGAAACAACUUUA------ACACCAGAAGGAAAUCC ....-------....((.-.(((....----...)))((((((....(((((((.(((((((....-))).))))....)))))))....))))))------.........))...... ( -25.10) >DroSim_CAF1 5568 91 + 1 UCUA-------CCAACCA-AUGGG--------------AGAGUCUAGUUGGCCUAGAUGAGGAAAUUCCUCCAUUACAGAGGCCGAAACAACUUUA------ACACCAGAAGGAAAUCC ....-------....((.-.(((.--------------(((((....(((((((.(((((((.....))).))))....)))))))....))))).------...)))...))...... ( -26.40) >DroEre_CAF1 6196 97 + 1 UC-----------AGCUA-ACGUGGAU----CCACCAAAAAGUAUGCUUGGCCUAUAUGAGGAAGUUCCGCCAUUGCAAGGGCCACUACAACUUUA------ACACUAAAGGAAAAUCC ..-----------(((..-((.(((..----...)))....))..)))((((((......((.....))((....))..))))))......(((((------....)))))........ ( -19.00) >DroAna_CAF1 6583 111 + 1 UCCAAGUCCUUGCCACUAAAAAUAGAUCGACUCACCCU--ACUCUGUCCAUCCUAGAUCAGGAAAUUGCUCUC------GGGCAGAGUCAACUUUUAUGGCUACACCAUAAGAAAAUCC ............................(((((..(((--..((((.......))))..)))....((((...------.)))))))))...((((((((.....))))))))...... ( -23.30) >consensus UCUA_______CCAACCA_AUGGGAAU____CCACCAUAGAGUCUAGUUGGCCUAGAUGAGGAAAUUCCUCCAUUACAAAGGCCGAAACAACUUUA______ACACCAGAAGAAAAUCC ......................................(((((.....((((((...(((((........)).)))...)))))).....)))))........................ ( -8.44 = -8.88 + 0.44)


| Location | 19,080,725 – 19,080,815 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -9.66 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19080725 90 + 23771897 AGUCUAGUUGGCCUAGAUGAGGAAAUUCCACUAUCACAAAGGCCGCAACAACUUUA------ACACCAGAAGAAAAUCCAGACAAAAGCAAUUUAA .((((.((.(((((.((((.((.....))..))))....))))))).....((((.------......)))).......))))............. ( -16.40) >DroSec_CAF1 5713 89 + 1 AGUCUAGUUGGCCUAGAUGAGGAAAU-CCUCCAUUACAAAGGCCGAAACAACUUUA------ACACCAGAAGGAAAUCCAGACAAAAGCAAUUUAA .((((..(((((((.(((((((....-))).))))....)))))))..........------.........((....))))))............. ( -21.80) >DroSim_CAF1 5586 90 + 1 AGUCUAGUUGGCCUAGAUGAGGAAAUUCCUCCAUUACAGAGGCCGAAACAACUUUA------ACACCAGAAGGAAAUCCAGACAAAAGCAAUUUAA .((((..(((((((.(((((((.....))).))))....)))))))..........------.........((....))))))............. ( -22.90) >DroEre_CAF1 6220 90 + 1 AGUAUGCUUGGCCUAUAUGAGGAAGUUCCGCCAUUGCAAGGGCCACUACAACUUUA------ACACUAAAGGAAAAUCCAGACAAAAGCAAUUUAA ....((((((((((......((.....))((....))..))))).......(((((------....)))))..............)))))...... ( -18.90) >DroAna_CAF1 6621 90 + 1 ACUCUGUCCAUCCUAGAUCAGGAAAUUGCUCUC------GGGCAGAGUCAACUUUUAUGGCUACACCAUAAGAAAAUCCAGACAAAAGCAAUUUAA (((((((((.((((.....))))....(....)------)))))))))....((((((((.....))))))))....................... ( -21.40) >consensus AGUCUAGUUGGCCUAGAUGAGGAAAUUCCUCCAUUACAAAGGCCGAAACAACUUUA______ACACCAGAAGAAAAUCCAGACAAAAGCAAUUUAA .((((...((((((...(((((........)).)))...))))))......((((.............)))).......))))............. ( -9.66 = -9.94 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:17 2006