
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,066,547 – 19,066,648 |
| Length | 101 |
| Max. P | 0.565024 |

| Location | 19,066,547 – 19,066,648 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -22.71 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19066547 101 + 23771897 UGUGUUGGAUCCUUGAGGCCGUAAAAUAGCCCACAAGGAG---------CAGAGGAGCAAAGGAGCCUGGCAGCAGGCUGGGCUCAAUUAGCUGUCAGUCGCAGGCCGAA (((((((..((((((.(((.........)))..))))))(---------(((..((......(((((..((.....))..)))))..))..))))))).))))....... ( -35.10) >DroSec_CAF1 20593 96 + 1 UGUGUUGGAUCCUUGGGGCCGUAAAAUAGCCCGCAAGGAG---------CAGAGGAGCAAAGGAG-----CAGCAGGCUGGGCUCAAUUAGCUGUCAGUCGCAGGCCGAA ....((((.((((((((((.........)))).))))))(---------(......((......)-----)....(((((((((.....))))..)))))))...)))). ( -31.60) >DroSim_CAF1 20805 93 + 1 UGUGUUGGAUCCUUGGGGCCGUAAAAUAGCCCGCAAGGAG---------CAGAGGAGCAAAGGAG--------CAGGCUGGGCUCAAUUAGCUGUCAGUCGCAGGCCGAA ....((((.((((((((((.........)))).))))))(---------(......((......)--------).(((((((((.....))))..)))))))...)))). ( -31.60) >DroEre_CAF1 22714 105 + 1 UGUGUUGGAUCCUUGGGGCCGUAAAAUAGCCCGCCAGGAGCCA-----GCAAAGGAGCAAAGGAGCUUGGGAGCAGGCUGAGCUCAAUUAGCUGUCAGUCGCAGGCCGAA ..((((((.((((.(((((.........)))).).)))).)))-----)))...((((......)))).((.((.(((((((((.....)))..))))))))...))... ( -36.90) >DroYak_CAF1 35806 110 + 1 UGUGUCGGAUCCUUGGGGCCGUAAAAUAGUCCGCAAGGAGCCAAAGGAGCAAGGGAGCAAAGGAGCUUGGGAGCAGGCUGGGCUCAAUUAGCUGUCAGUCGCAGGCCGAA ((((.(((.((((((((((.........)))).)))))).))...(((((((..((((.....((((((....))))))..))))..)).))).)).).))))....... ( -38.70) >DroAna_CAF1 8385 76 + 1 UGUGCAAGUUCCUUGGGGCCGUAAAAUAGCCCGUAAGGAG---------------------------------CAGGCUGAGCUCAAUUAGCUGUCAGACGCAGGACGA- (.(((..((((((((((((.........)))).)))))))---------------------------------).((((((......)))))).......))).)....- ( -27.30) >consensus UGUGUUGGAUCCUUGGGGCCGUAAAAUAGCCCGCAAGGAG_________CAGAGGAGCAAAGGAG______AGCAGGCUGGGCUCAAUUAGCUGUCAGUCGCAGGCCGAA ....((((.((((((((((.........)))).))))))....................................(((((((((.....))))..))))).....)))). (-22.71 = -23.27 + 0.56)

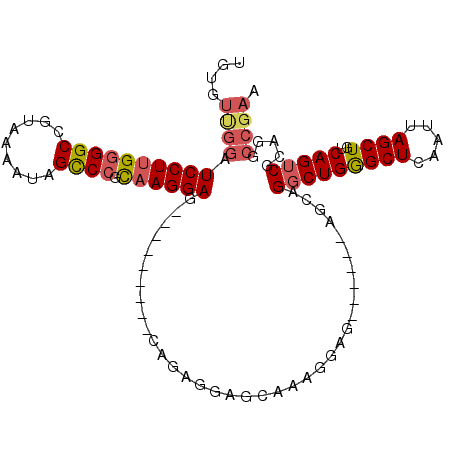
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:13 2006