| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,040,173 – 19,040,267 |
| Length | 94 |
| Max. P | 0.684338 |

| Location | 19,040,173 – 19,040,267 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.09 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -9.16 |
| Energy contribution | -10.55 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
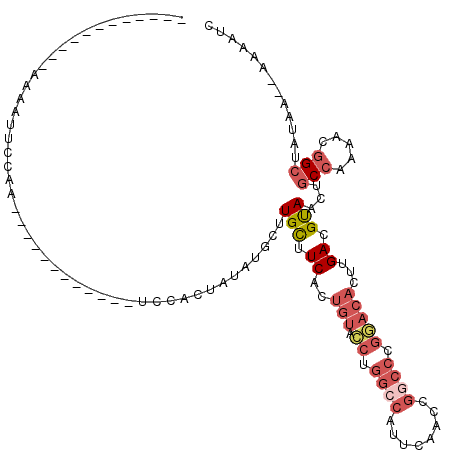
>3L_DroMel_CAF1 19040173 94 + 23771897 ------------AAACUUCAUA------------UCCACUACAUGCCUGCUUCACUGUACCUGGCCAUUCAACCGGCCCGGACACUUGACGUAACUGCCAAAACGGCUAUAA--AAAAUC ------------..........------------.............(((.(((.(((.((.((((........)))).)))))..))).)))...(((.....))).....--...... ( -19.10) >DroSec_CAF1 173437 94 + 1 ------------AAAAUUUCAA------------UCCACCAUAUGCAUGCUUCACUGUAUCUGGCCAUUCAACCGACCCGAACACUUGACGUAACUGCCAAAACGGCUAUAA--AAAAUC ------------..........------------....(((.(((((........))))).)))................................(((.....))).....--...... ( -9.80) >DroSim_CAF1 183114 94 + 1 ------------AAAAUUGCAA------------UCCACUAUAUGCUUGCUUCACUGUAUCUGGCCAUUCAACCGGCCCGAACACUUGACGUAACUGCCAAAACAGCUAUAA--AAAAUC ------------......(((.------------.........)))((((.(((.(((.((.((((........)))).)))))..))).))))(((......)))......--...... ( -14.80) >DroEre_CAF1 175556 106 + 1 UUGAUUAAACUGAAACUUCCAC------------UUCACUAUACGCUUGCUUCACUGUACCUGGCCAUUCAACCGGCCCGGACACUAGAAGUAACUGCCAA-ACGGCUUUAAA-AAAAUC ..........((((........------------))))........(((((((..(((.((.((((........)))).)))))...)))))))..(((..-..)))......-...... ( -22.80) >DroYak_CAF1 196960 119 + 1 AGCAUUUACUAUAAGCAUUUACUAUAUUUACAUAUACACUAUAUGCUUGCUUCACUGUACCUGGCCAUUCAACCGGCCCGGACACUAGACGUAACUGCCAA-ACGGCAUUAAAAAAAAUC .....((((..(((((((....(((((....)))))......)))))))......(((.((.((((........)))).)))))......)))).((((..-..))))............ ( -24.20) >DroAna_CAF1 170350 75 + 1 ------------AGAAUU--AUU-----------CUAAUUACAAGCUUGUUUCACUGAAACUGGGCAUUAAA------------CUUGACGCAACUACCAAACGGGCUUUAA-------- ------------......--...-----------........((((((((((..((......))((.(((..------------..))).)).......))))))))))...-------- ( -9.80) >consensus ____________AAAAUUCCAA____________UCCACUAUAUGCUUGCUUCACUGUACCUGGCCAUUCAACCGGCCCGGACACUUGACGUAACUGCCAAAACGGCUAUAA__AAAAUC ...............................................(((.((..(((.((.((((........)))).)))))...)).)))...(((.....)))............. ( -9.16 = -10.55 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:06 2006