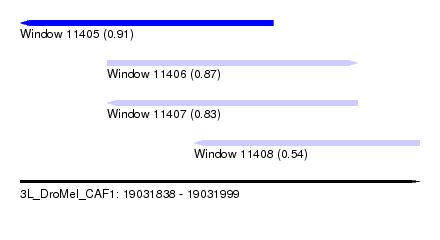
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,031,838 – 19,031,999 |
| Length | 161 |
| Max. P | 0.907683 |

| Location | 19,031,838 – 19,031,940 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -10.71 |
| Energy contribution | -12.47 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19031838 102 - 23771897 UGUUGUCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU--UUCCCUUUUGGUUUUUGUUAGCGUAAGAAGAUACAAACCAUUUUACAUAU ...((((((-(-((((((.....)))...))))))))))...............--........((((((.((((..(....)..)))).)))))).......... ( -30.90) >DroSec_CAF1 165587 99 - 1 UGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCCAAACUGCUGAGAUUU--UUCCCUUUUGGUUUUUGUUAGCGUAA---GAUACAAACCAUUUUACAUAU .((((((((-(-((((((.....)))...)))))))))..))).(((((.(...--..((.....))....).)))))((((---(((.......))))))).... ( -32.60) >DroSim_CAF1 174671 99 - 1 UGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU--UUCCCUUUUGGUUUUUGUUAGCGUAA---GAUACAAACCAUUUUACAUAU ...((((((-(-((((((.....)))...)))))))))).....(((((.(...--..((.....))....).)))))((((---(((.......))))))).... ( -34.00) >DroEre_CAF1 166881 99 - 1 UGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCGUGGGAAUAACUGCUGAGAUUUGUUUCCUUUUUGGUUUUUGUUAGCGUAA---GAU--AAAAGAUUUUACAUAU ((((.((((-(-((((((.....)))...))))))))))))...(((((.(.......((.....))....).)))))((((---(((--.....))))))).... ( -30.60) >DroYak_CAF1 188344 99 - 1 UGUUGCCCA-CGGCCCCCGAAAUGUGCGUGCCAUGGGCAUAACUGCUGAGAUUU-UUUCCUUUUUGGUUUUUGUUAGCAUUA---GAU--AAACAAUUUUACAUAU (((((((((-.(((.(.((.....)).).))).))))).....((((((.(...-...((.....))....).))))))...---...--.))))........... ( -23.30) >DroMoj_CAF1 190192 80 - 1 UGUUAUUGAACGGCCCUCGCA---GCCGAGGCGAGGGCGUAAAUU---AG-UUU--UUACAUUUAAG------------U-----GAAACACACAAUUUUCCUUAU ............((((((((.---......))))))))(((((..---..-..)--))))...((((------------.-----((((........)))))))). ( -23.40) >consensus UGUUGCCCA_U_GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU__UUCCCUUUUGGUUUUUGUUAGCGUAA___GAUACAAACCAUUUUACAUAU ...((((((...((((((.....)))...))).)))))).....(((((.(.......((.....))....).)))))............................ (-10.71 = -12.47 + 1.75)

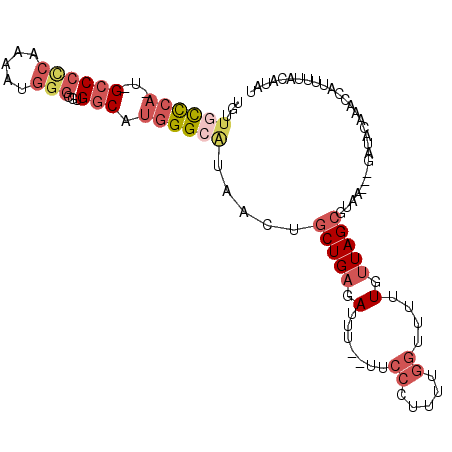
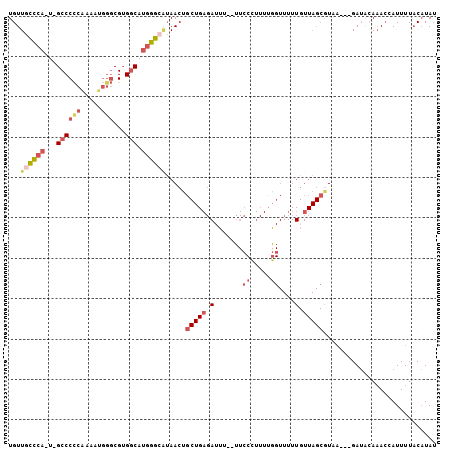
| Location | 19,031,873 – 19,031,974 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -18.79 |
| Energy contribution | -21.52 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19031873 101 + 23771897 AAAACCAAAAGGGAA--AAAUCUCAGCAGUUAUGCCCAUGCCACGCCCAUUUUGGGGGC-A-UGGACAACAAAUGCGCUGCCUGGCAGCAAAAUUUCG-AUUCCAU ...........((((--........(((....((.(((((((...(((.....))))))-)-))).)).....)))(((((...))))).........-.)))).. ( -32.40) >DroSec_CAF1 165619 101 + 1 AAAACCAAAAGGGAA--AAAUCUCAGCAGUUUGGCCCAUGCCACGCCCAUUUUGGGGGC-A-UGGGCAACAAAUGCGCUGCCUGGCAGCAAAAUUUCG-AUUCCAU ...........((((--........(((.(((((((((((((...(((.....))))))-)-)))))..)))))))(((((...))))).........-.)))).. ( -39.00) >DroSim_CAF1 174703 101 + 1 AAAACCAAAAGGGAA--AAAUCUCAGCAGUUAUGCCCAUGCCACGCCCAUUUUGGGGGC-A-UGGGCAACAAAUGCGCUGCCUGGCAGCAAAAUUUCG-AUUCCAU ...........((((--........(((....((((((((((...(((.....))))))-)-)))))).....)))(((((...))))).........-.)))).. ( -39.50) >DroEre_CAF1 166911 103 + 1 AAAACCAAAAAGGAAACAAAUCUCAGCAGUUAUUCCCACGCCACGCCCAUUUUGGGGGC-A-UGGGCAACAAACGCGCUGCCUGGCAGCCAAAUUUCG-AUUCCAU ...........((((.(........((.(((...((((.(((...(((.....))))))-.-)))).....)))))(((((...)))))........)-.)))).. ( -28.10) >DroYak_CAF1 188374 103 + 1 AAAACCAAAAAGGAAA-AAAUCUCAGCAGUUAUGCCCAUGGCACGCACAUUUCGGGGGCCG-UGGGCAACACAUGCGCUGCCUGGCAGCAAAAUUUCG-AUUCCAU ...........((((.-........(((((..((((((((((.(.(........).)))))-))))))..)).)))(((((...))))).........-.)))).. ( -37.00) >DroMoj_CAF1 190214 92 + 1 ----CUUAAAUGUAA--AAA-CU---AAUUUACGCCCUCGCCUCGGC---UGCGAGGGCCGUUCAAUAACAAUUCUGCUGCCUGGCAGCCAA-UUUCGUUUUCCAU ----.......((((--(..-..---..)))))((((((((......---.)))))))).................(((((...)))))...-............. ( -24.30) >consensus AAAACCAAAAGGGAA__AAAUCUCAGCAGUUAUGCCCAUGCCACGCCCAUUUUGGGGGC_A_UGGGCAACAAAUGCGCUGCCUGGCAGCAAAAUUUCG_AUUCCAU ...........((((..........(((....((((((.(((...(((.....))))))...)))))).....)))(((((...)))))...........)))).. (-18.79 = -21.52 + 2.72)

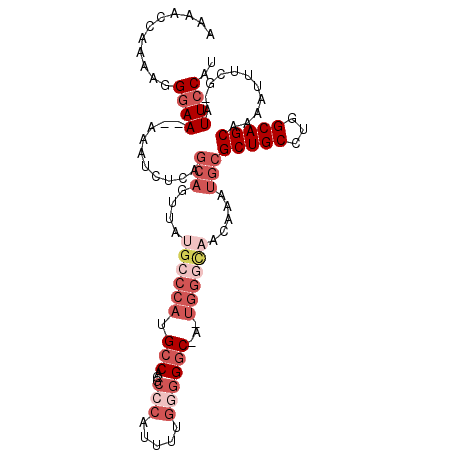
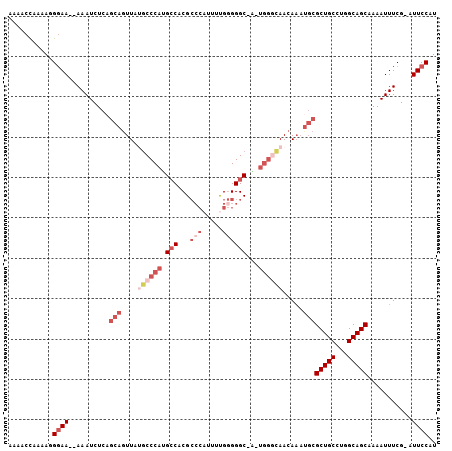
| Location | 19,031,873 – 19,031,974 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -22.50 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19031873 101 - 23771897 AUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGUCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU--UUCCCUUUUGGUUUU ..(((..-.(((((((((((((...)))))(((.((((.(((((-(-((((((.....)))...))))))))))))).))).)))))))--))))........... ( -38.30) >DroSec_CAF1 165619 101 - 1 AUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCCAAACUGCUGAGAUUU--UUCCCUUUUGGUUUU ..(((..-.(((((((((((((...)))))(((((((..(((((-(-((((((.....)))...))))))))))))).))).)))))))--))))........... ( -39.50) >DroSim_CAF1 174703 101 - 1 AUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU--UUCCCUUUUGGUUUU ..(((..-.(((((((((((((...)))))(((.((((.(((((-(-((((((.....)))...))))))))))))).))).)))))))--))))........... ( -41.00) >DroEre_CAF1 166911 103 - 1 AUGGAAU-CGAAAUUUGGCUGCCAGGCAGCGCGUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCGUGGGAAUAACUGCUGAGAUUUGUUUCCUUUUUGGUUUU ..((((.-((((.(((.(((((...)))))(((.(((((.((((-(-((((((.....)))...))))))))))))).))).))).)))).))))........... ( -38.60) >DroYak_CAF1 188374 103 - 1 AUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUGUGUUGCCCA-CGGCCCCCGAAAUGUGCGUGCCAUGGGCAUAACUGCUGAGAUUU-UUUCCUUUUUGGUUUU ..((((.-.(((((((((((((...)))))(((.((..((((((-.(((.(.((.....)).).))).))))))..))))).)))))))-)))))........... ( -34.20) >DroMoj_CAF1 190214 92 - 1 AUGGAAAACGAAA-UUGGCUGCCAGGCAGCAGAAUUGUUAUUGAACGGCCCUCGCA---GCCGAGGCGAGGGCGUAAAUU---AG-UUU--UUACAUUUAAG---- ......(((((..-...(((((...)))))....))))).(((((..((((((((.---......))))))))(((((..---..-..)--)))).))))).---- ( -30.00) >consensus AUGGAAU_CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA_U_GCCCCCAAAAUGGGCGUGGCAUGGGCAUAACUGCUGAGAUUU__UUCCCUUUUGGUUUU ..((((....((((((((((((...)))))(((.((((.(((((...((((((.....)))...))).))))))))).))).)))))))..))))........... (-22.50 = -24.05 + 1.55)
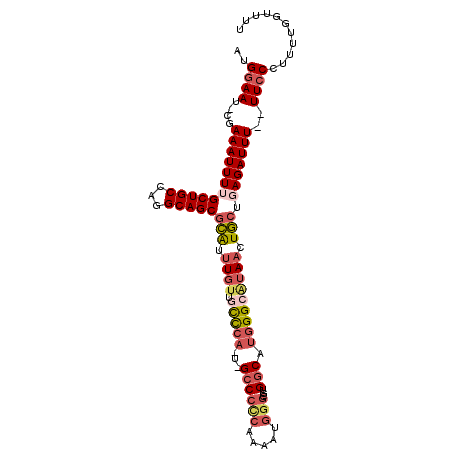
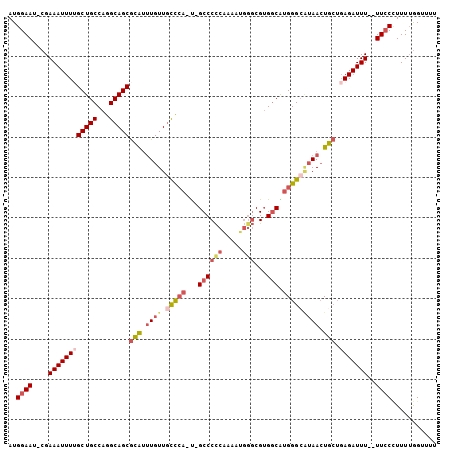
| Location | 19,031,908 – 19,031,999 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.98 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19031908 91 - 23771897 UCCACUUUUUACAAGCUAAUGCGAAAUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGUCCA-U-GCCCCCAAAAUGGGCGUGGCAU .............(((.((((((((((.....-....)))))(((((...)))))))))).)))(.(((-(-((((.......))))))))).. ( -28.50) >DroSec_CAF1 165654 91 - 1 UCCACUUUUUACAAGCUAAUGCGAAAUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAU .((((........(((.((((((((((.....-....)))))(((((...)))))))))).)))(((((-(-.........))))))))))... ( -29.00) >DroSim_CAF1 174738 91 - 1 UCCACUUUUUACACGCUAAUGCGAAAUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCAU ....................(((((((.....-....)))))))((((.(((((((....))))))).(-(-((((.......)))))))))). ( -28.40) >DroEre_CAF1 166948 91 - 1 UUCACUUUUUACAAGCUAAUGCGAAAUGGAAU-CGAAAUUUGGCUGCCAGGCAGCGCGUUUGUUGCCCA-U-GCCCCCAAAAUGGGCGUGGCGU .............((((((..(((.......)-))....))))))(((.(((((((....))))))).(-(-((((.......))))))))).. ( -29.90) >DroYak_CAF1 188410 92 - 1 UCCACUUUUUACAAGCUAAUGCGAAAUGGAAU-CGAAAUUUUGCUGCCAGGCAGCGCAUGUGUUGCCCA-CGGCCCCCGAAAUGUGCGUGCCAU ..............((....))...((((...-((..((((((..(((.(((((((....)))))))..-.)))...))))))...))..)))) ( -25.60) >DroMoj_CAF1 190241 81 - 1 ------UUGUACAUGGAAA---AAUAUGGAAAACGAAA-UUGGCUGCCAGGCAGCAGAAUUGUUAUUGAACGGCCCUCGCA---GCCGAGGCGA ------.....((((....---..)))).....((...-((((((((..(((..(((........)))....)))...)))---)))))..)). ( -22.20) >consensus UCCACUUUUUACAAGCUAAUGCGAAAUGGAAU_CGAAAUUUUGCUGCCAGGCAGCGCAUUUGUUGCCCA_U_GCCCCCAAAAUGGGCGUGGCAU ....................(((((((..........)))))))((((((((((((....))))))).....((((.......)))).))))). (-17.70 = -18.98 + 1.28)

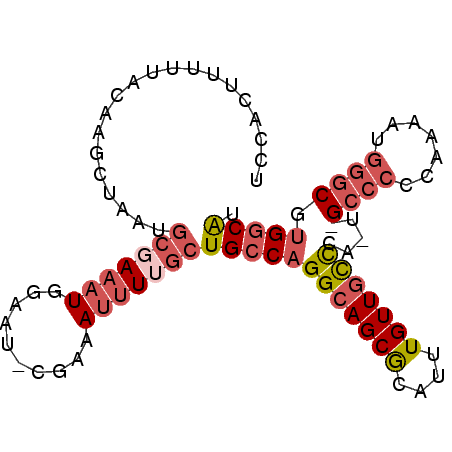
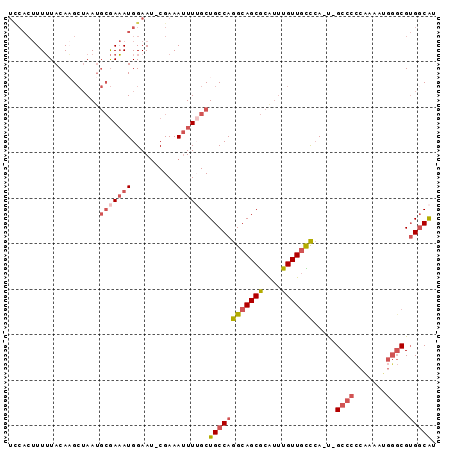
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:02 2006