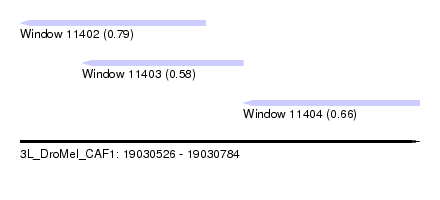
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,030,526 – 19,030,784 |
| Length | 258 |
| Max. P | 0.786673 |

| Location | 19,030,526 – 19,030,646 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19030526 120 - 23771897 UUACAAGCACCCAACCAAAAGGGUCUGGAAUGGAGCAUUAAUCCACUUGAUGCGCCUGUAGGCAGGCAGUUUCUCCAGCCUCUCCAUUAUCAAUGGCAGCACCAGCAUUCCGGGCGCCGC ......((.(((..((....))(.((((..(((((((((((.....)))))))(((((....)))))......))))(((..............)))....))))).....))).))... ( -39.44) >DroVir_CAF1 178892 120 - 1 UUACAAGCAGCCCACCAUGAGCGUUUGGAAGGGUGCAUUGAUCCAUUUGAUGCGCCUGUAUGCGGGCAACUUCUCCAGUCGCUGCAUAAUGACGGGCAGCAACAACAUGCCCGGCGCUGC ......(((((((...(((((((.(((((..(((((((..(.....)..))))(((((....))))).)))..))))).)))).)))......(((((.........))))))).))))) ( -53.60) >DroGri_CAF1 168129 120 - 1 UUACAAGCAGCCCACCAUCAGUGUUUGGAAUGGCGCAUUGAUCCAUUUGAUGCGUGCAUAGGCGGGCAGCUUCUCCAGCCUCUGCAUAAUCACUGGCAGCACCAGCAUGCCGGGUGCAGC ......(((.(((......((((.(((.....((((((..(.....)..))))))(((.((((.((.((...)))).)))).))).))).))))((((((....)).))))))))))... ( -44.40) >DroWil_CAF1 209789 120 - 1 UUACAAGCAGCCAACCAUUAGAGUCUGGAAUGGUGCAUUUGCCCAUUUGAUUUUCUUAUACACUGGGAAUUUCUCCAAUUUCUCCAUGAUCAAUGGCAACACCAACAUGCCAGGUGCUGC ......(((((..(((...((((..((((((((.((....))))))..((..((((((.....))))))..))))))..))))..........(((((.........))))))))))))) ( -33.20) >DroMoj_CAF1 188828 120 - 1 UCACAAGCAGCCCACCAUGAGCGUUUGGAACGGCGCAUUGAUCCACUUGAUGCGUUUGUACGCUGGCAGUUUCUCCAGUCGCUCCAUGACAACGGGCAGUAGCAGCAUACCCGGGGCUGC ......(((((((..((((((((.(((((...((((((..(.....)..))))))((((......))))....))))).)))).))))....((((..((....))...))))))))))) ( -46.90) >DroPer_CAF1 172645 120 - 1 UUACAAGCAUCCAACCAUCAGGGUUUGGAAAGGGGCAUUUAUCCAUUUGAUGCGGCUGUAGGCGGGCAGCUUUUCCAAUCGCUCCAUGAUCAAAGGCAGUACCAGCAUCCCGGGCGCUGC .....(((..((....(((((((((((((((((.(((((.........)))))(.(((....))).)..)))))))))..)).)).))))....((.....))........))..))).. ( -31.60) >consensus UUACAAGCAGCCAACCAUCAGCGUUUGGAAUGGCGCAUUGAUCCAUUUGAUGCGCCUGUAGGCGGGCAGCUUCUCCAGUCGCUCCAUGAUCAAUGGCAGCACCAGCAUGCCGGGCGCUGC ......(((((...((...((((.(((((..((((((((((.....)))))))(((((....))))).)))..))))).))))..........(((.............))))).))))) (-24.34 = -24.90 + 0.56)

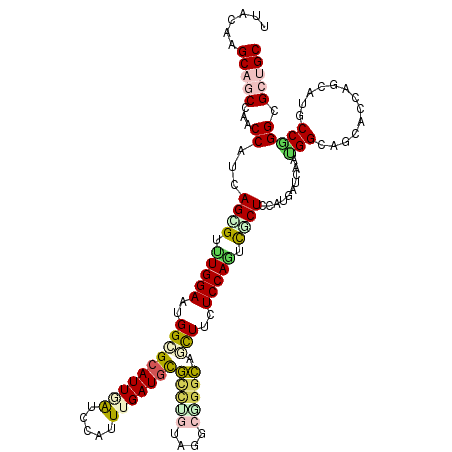
| Location | 19,030,566 – 19,030,670 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19030566 104 - 23771897 AGUGGUCGUAG--AUGAUAA--AGUUACUUACAAGCACCCAACCAAAAGGGUCUGGAAUGGAGCAUUAAUCCACUUGAUGCGCCUGUAGGCAGGCAGUUUCUCCAGCC .(((.(.((((--.((((..--.)))).)))).).)))((((((.....))).)))..(((((((((((.....)))))))(((((....)))))......))))... ( -30.40) >DroSec_CAF1 164289 104 - 1 AGUGCUCGUAG--ACGGUAA--AGUUACUUACAAGCACCCAACCAAUAGGGUCUGGAAUGGAGCAUUGAUCCACUUGAUGCGCCUGUACGCAGGCAGUUUCUCCAGUU .(((((.((((--((.....--.)))...))).)))))((((((.....))).)))..((((((((..(.....)..))))(((((....)))))......))))... ( -34.30) >DroSim_CAF1 173360 102 - 1 AGUGGUGGUAG--AUGAU----AGUUACUUACAAGCACCCUACCAAUAAGGUCUGGAAUGGAGCAUUGAUCCACUUGAUGCGCCUGUACGCAGGCAGUUUCUCCAGCC .....((((((--.((..----.((.....))...))..))))))....((.(((((.....((((..(.....)..))))(((((....)))))......))))))) ( -32.00) >DroYak_CAF1 187004 101 - 1 AGUGGUCG-----AGGGUAA--AUUUACUUACAAGCACCCAACCAAUAGGGUCUGGAAUGGAGCAUUAAUCCACUUGAUGCGCCUGUAGGCAGGCAGUUUCUCCAGCC ..((((..-----.((((..--..............)))).))))....((.(((((.....(((((((.....)))))))(((((....)))))......))))))) ( -31.29) >DroAna_CAF1 161408 101 - 1 -UU--ACGUAGUUAUCUU----AACGACUUACAAGCACCCAACCAUCAGGGUCUGGAAGGGAGCAUUUAUCCACUUGAUGCGCCUGUAGGCGGGCAGCUUCUCCAGCC -..--..((((((.....----...))).)))....((((........))))(((((..((.(((((.........)))))(((((....)))))..))..))))).. ( -29.00) >DroPer_CAF1 172685 106 - 1 AUUGCUUAUAG--GUGAUCGUGAUCCCCUUACAAGCAUCCAACCAUCAGGGUUUGGAAAGGGGCAUUUAUCCAUUUGAUGCGGCUGUAGGCGGGCAGCUUUUCCAAUC ..(((((..((--(.(((....))).)))...)))))(((((((.....)).)))))..((((((((.........)))))((((((......))))))..))).... ( -30.50) >consensus AGUGGUCGUAG__AUGAUAA__AGUUACUUACAAGCACCCAACCAAUAGGGUCUGGAAUGGAGCAUUAAUCCACUUGAUGCGCCUGUAGGCAGGCAGUUUCUCCAGCC ....................................((((........))))(((((..((.(((((((.....)))))))(((((....)))))..))..))))).. (-23.32 = -23.02 + -0.30)

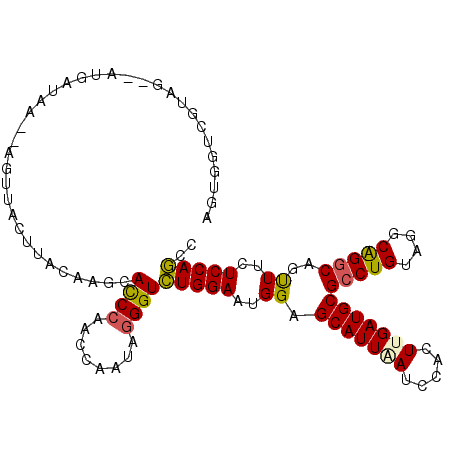
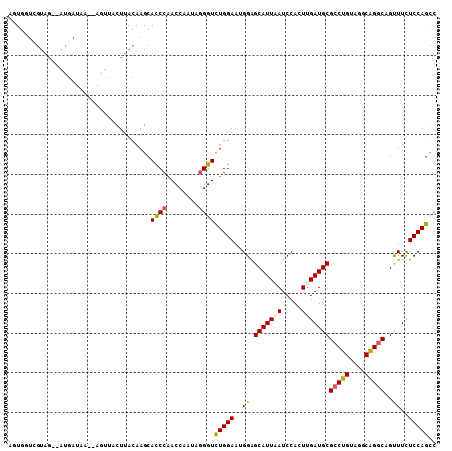
| Location | 19,030,670 – 19,030,784 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.65 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19030670 114 - 23771897 GGUGCGCAUAAUCGAAGUGUCCAAGGAACACUGCUGUGGGAACAGGGCGCAGGCAGUGGGCACCAUGAAGCAAAGGCUAAACAUCAAAGA---AUAAAAAGAUCAGUUAAAGGAACA (((((.(((..((..((((((....).)))))(((.((....)).)))...))..))).)))))....(((....)))............---........................ ( -29.00) >DroSec_CAF1 164393 114 - 1 GGUGCGCAUAAUCGAAGUGUCCAAGGAACACUGCUGUGGGAACAGGGCGCAGGCAGUGGGCACCAUGAAGCAAAGGCUAAACAUCAAAGA---AUUGCAAGAUUAGCUAAAGGAACU (((((.(((..((..((((((....).)))))(((.((....)).)))...))..))).)))))..........((((((.(..(((...---.)))...).))))))......... ( -31.80) >DroSim_CAF1 173462 114 - 1 GGUGCGCAUAAUCGAAGUGUCCAAGGAACACUGCUGUGGGAACAGGGCGCAGGCAGUGGGCACCAUGAAGCAAAGGCUAAACAUCAAAUA---AGUGCAAGAUUAGCUAAAGGAUCU ..(((((....(((..(((((((..(.....((((.((....)).))))....)..)))))))..)))(((....)))............---.)))))(((((........))))) ( -32.30) >DroEre_CAF1 165631 113 - 1 GGUGCGCAUAAUCGAAGUGUCCAAGGAACACUGCUGUGGAAACAGGGCGCAGGCAGUGGGCACCAUGAAGCAAAGGCUAAA-GUUAACGA---AUAGAUAGAUUAGCUGAAGGAACU (((((.(((..((..((((((....).)))))(((.((....)).)))...))..))).)))))..........((((((.-.(((.(..---...).))).))))))......... ( -33.30) >DroYak_CAF1 187105 114 - 1 GGUGCGCAUAAUCGAAGUGUCCAAAGAACACUGCUGGGGAAACAGGGCGCAGGCAGUGGGCACCAUAAAGCAGAGGCUAAACAUUAAAGA---AAAGAUAUAUUAGCUGAAAGAACU (((((.(((..((..(((((.......)))))(((..(....)..)))...))..))).)))))..........((((((..(((.....---...)))...))))))......... ( -28.10) >DroAna_CAF1 161509 114 - 1 GGUGCGCAUGGUGGACGUGUCCAAGGAGCAUUGUUGCGGGAACAGAGCGCAGGCAGAGGGCACCAUGAAGCAGAGGCUGAAA-UUGAUAAGAAAUUUUAUUAUUA--UUGAGGAAUA ..(((.(((((((..(.((((....(.((.(((((.....))))).)).).))))..)..)))))))..)))....((.((.-.(((((((....)))))))...--)).))..... ( -26.50) >consensus GGUGCGCAUAAUCGAAGUGUCCAAGGAACACUGCUGUGGGAACAGGGCGCAGGCAGUGGGCACCAUGAAGCAAAGGCUAAACAUCAAAGA___AUAGAAAGAUUAGCUAAAGGAACU (((((.(((......((((((....).)))))(((.((....)).))).......))).)))))....(((....)))....................................... (-25.54 = -25.65 + 0.11)

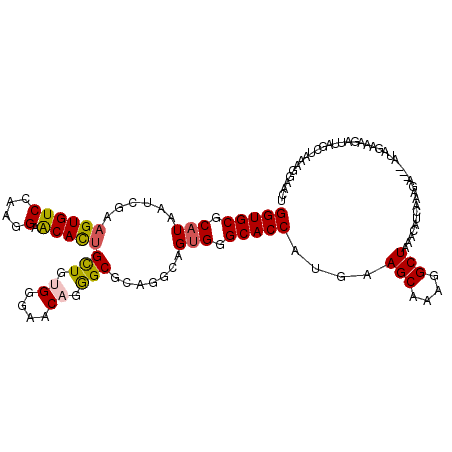

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:58 2006