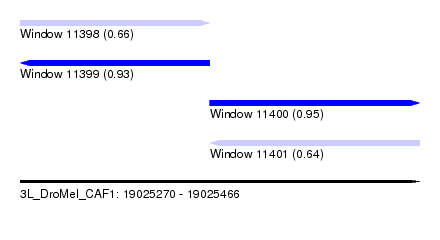
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,025,270 – 19,025,466 |
| Length | 196 |
| Max. P | 0.951881 |

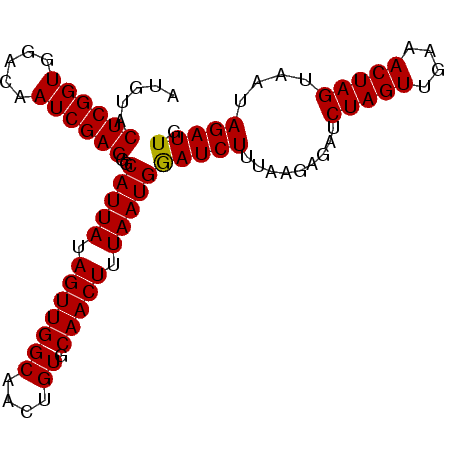
| Location | 19,025,270 – 19,025,363 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.59 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19025270 93 + 23771897 AUGUACUCGGUGGACAAUCGAGGGCAUUAUAGUUGGCAACUGUGCAACUUUAAUGGAUCUUUAAGAGAUCUAGUUGAAACUAGUAAUAGAUUG (((..((((((.....))))))..)))((((((.....))))))(((((.....(((((((...))))))))))))...(((....))).... ( -23.70) >DroSec_CAF1 159068 93 + 1 AUGUACUCGGUGGACAAUCGAGGGCAUUAUAGUUGGCAACUGUGCAACUUUAAUGAAUCUUUAAGAGAUCUAGUUGAAACUAGUAAUAGAUUG .....((((((.....))))))..(((((.(((((((....)).))))).)))))(((((.........(((((....)))))....))))). ( -21.22) >DroSim_CAF1 167685 93 + 1 AUGUACUCGGUGGACAAUCGAGGUCAUUAUAGUUGGCAACUGUGCAACUUUAAUGGAUCUUUAAGAGAUCUAGUUGAAACUAGUAAUAGAUUG .....((((((.....))))))..(((((.(((((((....)).))))).)))))((((((...)))))).........(((....))).... ( -22.70) >consensus AUGUACUCGGUGGACAAUCGAGGGCAUUAUAGUUGGCAACUGUGCAACUUUAAUGGAUCUUUAAGAGAUCUAGUUGAAACUAGUAAUAGAUUG .....((((((.....))))))..(((((.(((((((....)).))))).)))))(((((.........(((((....)))))....))))). (-21.81 = -21.59 + -0.22)

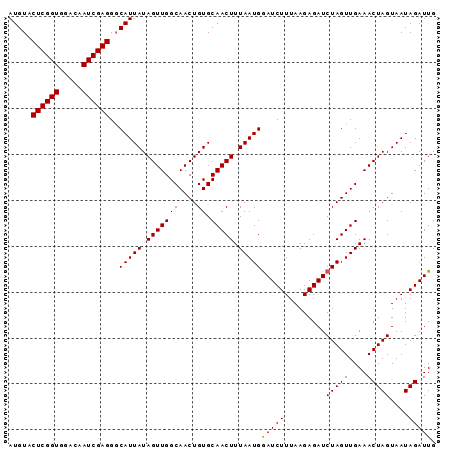
| Location | 19,025,270 – 19,025,363 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -16.63 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19025270 93 - 23771897 CAAUCUAUUACUAGUUUCAACUAGAUCUCUUAAAGAUCCAUUAAAGUUGCACAGUUGCCAACUAUAAUGCCCUCGAUUGUCCACCGAGUACAU (((((.....(((((....)))))..........((..(((((.(((((((....)).))))).)))))...))))))).............. ( -16.60) >DroSec_CAF1 159068 93 - 1 CAAUCUAUUACUAGUUUCAACUAGAUCUCUUAAAGAUUCAUUAAAGUUGCACAGUUGCCAACUAUAAUGCCCUCGAUUGUCCACCGAGUACAU (((((.....(((((....)))))..........((..(((((.(((((((....)).))))).)))))...))))))).............. ( -16.60) >DroSim_CAF1 167685 93 - 1 CAAUCUAUUACUAGUUUCAACUAGAUCUCUUAAAGAUCCAUUAAAGUUGCACAGUUGCCAACUAUAAUGACCUCGAUUGUCCACCGAGUACAU (((((.....(((((....)))))..............(((((.(((((((....)).))))).))))).....))))).............. ( -16.70) >consensus CAAUCUAUUACUAGUUUCAACUAGAUCUCUUAAAGAUCCAUUAAAGUUGCACAGUUGCCAACUAUAAUGCCCUCGAUUGUCCACCGAGUACAU (((((.....(((((....)))))..........((..(((((.(((((((....)).))))).)))))...))))))).............. (-16.60 = -16.60 + -0.00)

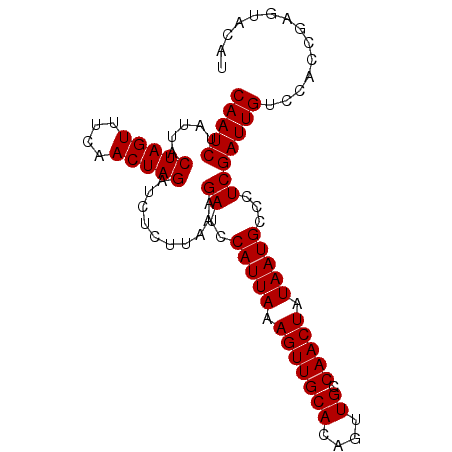
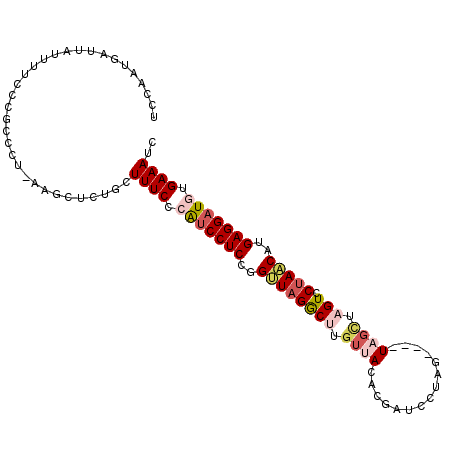
| Location | 19,025,363 – 19,025,466 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -23.11 |
| Consensus MFE | -14.41 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19025363 103 + 23771897 ACCAAUGAUUAUUUUCCCGCUCU-AAGCUCUGCUUUCCCAUCCUCCGGCU-GGCUUGUUAGACGAUCUUACAGUUUUGCUAGUCCUAACAUGAGGAUGUGAAACC ..................((...-..)).....((((.(((((((.((((-(((((((.((.....)).))))....))))))).......))))))).)))).. ( -23.10) >DroSec_CAF1 159161 99 + 1 UCCAAUGAUUAUUUUCCCGCCCU-AAGCUCUGCUUUCCCAUCCUCCGGUUAGACUUGUUACAC-AUCUUAG----UAGCUAGUCCUAACAUGAGGAUGUGAAAUC ..................((...-..)).....((((.(((((((..((((((((.(((((..-......)----)))).))).)))))..))))))).)))).. ( -27.10) >DroSim_CAF1 167778 99 + 1 UCCAAUGAUUAUUUUCCCGCCCU-AAGCUCUGCUUUCCCAUCCUCCCGUUAGGCUUGUUACAC-AUCCUAG----UAGCUAGUCCUAACAUGAGGAUGUGAAAUC ..................((...-..)).....((((.(((((((..((((((((.(((((..-......)----)))).)).))))))..))))))).)))).. ( -28.80) >DroEre_CAF1 160198 104 + 1 UCCAGUGAUUUAUUUCCCACCCU-AAGCCCUGCUUUCCCGUCCUCUUGCUAGACCAAUUAUACGAUACUGAAGUUUAGCUAGUCCUAGCAUGAGGAUUUGAAAUC ....(((..........)))...-.........((((..((((((.((((((((...(((.((.........)).)))...)).)))))).))))))..)))).. ( -20.60) >DroYak_CAF1 181734 93 + 1 UCCAGUGAAUAAAUCCCCACCCCAAAGCCCUGCUUUCCCGUCCUCUUGUUAGGCCAGUU-UACGACC-----------UUAGUCCUAACAUGAGGUUAUGAAAUC ....(((..........))).............((((....((((.(((((((.(....-.......-----------...).))))))).))))....)))).. ( -15.94) >consensus UCCAAUGAUUAUUUUCCCGCCCU_AAGCUCUGCUUUCCCAUCCUCCGGUUAGGCUUGUUACACGAUCCUAG____UAGCUAGUCCUAACAUGAGGAUGUGAAAUC .................................((((.(((((((..((((((((.((((...............)))).))).)))))..))))))).)))).. (-14.41 = -15.82 + 1.41)


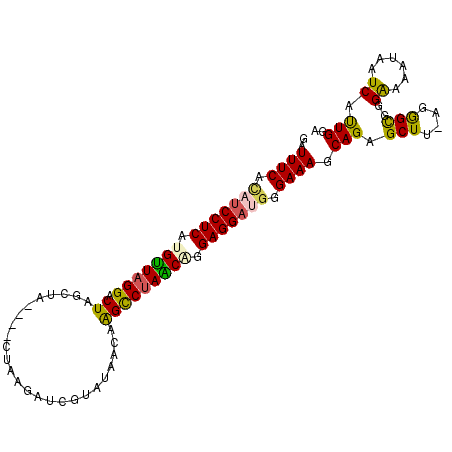
| Location | 19,025,363 – 19,025,466 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -21.07 |
| Energy contribution | -20.59 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19025363 103 - 23771897 GGUUUCACAUCCUCAUGUUAGGACUAGCAAAACUGUAAGAUCGUCUAACAAGCC-AGCCGGAGGAUGGGAAAGCAGAGCUU-AGAGCGGGAAAAUAAUCAUUGGU (.((((.(((((((.(((((((.((.(((....))).))....))))))).(..-..)..))))))).)))).)...((..-...)).................. ( -23.90) >DroSec_CAF1 159161 99 - 1 GAUUUCACAUCCUCAUGUUAGGACUAGCUA----CUAAGAU-GUGUAACAAGUCUAACCGGAGGAUGGGAAAGCAGAGCUU-AGGGCGGGAAAAUAAUCAUUGGA ..((((.(((((((..(((((.(((.(.((----(......-..))).).))))))))..))))))).)))).....((..-...)).................. ( -25.10) >DroSim_CAF1 167778 99 - 1 GAUUUCACAUCCUCAUGUUAGGACUAGCUA----CUAGGAU-GUGUAACAAGCCUAACGGGAGGAUGGGAAAGCAGAGCUU-AGGGCGGGAAAAUAAUCAUUGGA ..((((.(((((((.(((((((.((.(.((----(......-..))).).))))))))).))))))).)))).....((..-...)).................. ( -28.80) >DroEre_CAF1 160198 104 - 1 GAUUUCAAAUCCUCAUGCUAGGACUAGCUAAACUUCAGUAUCGUAUAAUUGGUCUAGCAAGAGGACGGGAAAGCAGGGCUU-AGGGUGGGAAAUAAAUCACUGGA ..((((...(((((.((((((.(((((.((.((.........)).)).))))))))))).)))))...)))).........-..(((((........)))))... ( -26.70) >DroYak_CAF1 181734 93 - 1 GAUUUCAUAACCUCAUGUUAGGACUAA-----------GGUCGUA-AACUGGCCUAACAAGAGGACGGGAAAGCAGGGCUUUGGGGUGGGGAUUUAUUCACUGGA ..........((((.(((((((.(((.-----------(......-..))))))))))).)))).....(((((...)))))..((((((......))))))... ( -24.70) >consensus GAUUUCACAUCCUCAUGUUAGGACUAGCUA____CUAAGAUCGUAUAACAAGCCUAACAGGAGGAUGGGAAAGCAGAGCUU_AGGGCGGGAAAAUAAUCAUUGGA ..((((.(((((((.(((((((.((.........................))))))))).))))))).)))).(((.(((....)))..((......)).))).. (-21.07 = -20.59 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:56 2006