| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,023,380 – 19,023,477 |
| Length | 97 |
| Max. P | 0.890720 |

| Location | 19,023,380 – 19,023,477 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
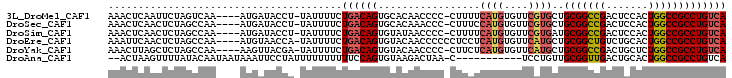
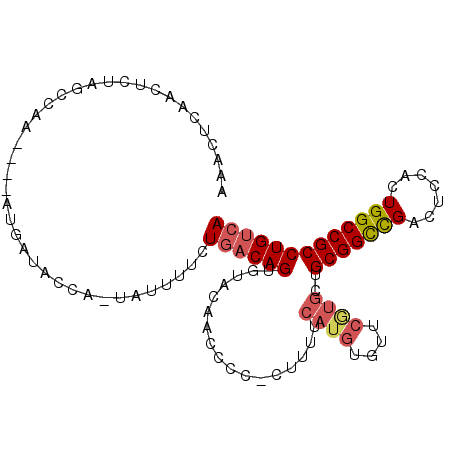
>3L_DroMel_CAF1 19023380 97 + 23771897 AAACUCAAUUCUAGUCAA----AUGAUACCU-UAUUUUCUGACAGUGCACAACCCC-CUUUUCAUGUGUUCGUGCUGCGGCCGACUCCACUGGCCGCCUGUCA .............(((..----..)))....-........(((((.(((((((...-..........))).)))).(((((((.......)))))))))))). ( -24.22) >DroSec_CAF1 157197 97 + 1 AAACUCAACUCUAGCCAA----AUGAUACCU-UAUUUUCUGACAGUGCACAAACCC-CUUUCCAUGUGUUCGUGCUGCGGCCGACUCCACUGGCCGCCUGUCA ..................----.........-........(((((.((((.(((.(-........).))).)))).(((((((.......)))))))))))). ( -25.00) >DroSim_CAF1 165799 97 + 1 AAACUCAACUCUAGCCAA----AUGAUACCU-UAUUUUCUGACAGUGUAUAACCCC-CUUUUCAUGUGUUCGUGAUGCGGCCGACUCCACUGGCCGCCUGUCA ..................----.........-........(((((...........-....(((((....))))).(((((((.......)))))))))))). ( -22.90) >DroEre_CAF1 158121 98 + 1 AAAUUCAACUCUAGCCAA----AUGUAACCA-UAUUUUCUGACAGUGUACAACCCCCCUCCUCAUGUGUUCAUGCUGCGGCUGUCUGCACUGGCCGCCUGUCA ..................----.........-........(((((.................((((....))))..((((((.........))))))))))). ( -18.20) >DroYak_CAF1 179823 97 + 1 AAACUUAGCUCUAGCCAA----AAGUUACGA-UAUUUUCUGACAGUGUACAACCCC-CUUCUCAUGUGUUCAUGCUGCGGCCGACUGCUCUGGCCGCCUGUCA .(((((.((....))...----)))))....-........(((((...........-.....((((....))))..(((((((.......)))))))))))). ( -23.80) >DroAna_CAF1 154801 89 + 1 --ACUAAGUUUUAUACAAUAAUAAAUUCCUAUUUUUUUUUUCCAGUGUAAGACUAA-C-----------UCCUGUUGCGGUUGACUGCACUGGCCGCCUGUCA --....((((((((((............................))))))))))..-.-----------.......((((((.........))))))...... ( -14.59) >consensus AAACUCAACUCUAGCCAA____AUGAUACCA_UAUUUUCUGACAGUGUACAACCCC_CUUUUCAUGUGUUCGUGCUGCGGCCGACUCCACUGGCCGCCUGUCA .......................................((((((.................((((....))))..(((((((.......))))))))))))) (-16.76 = -17.23 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:52 2006