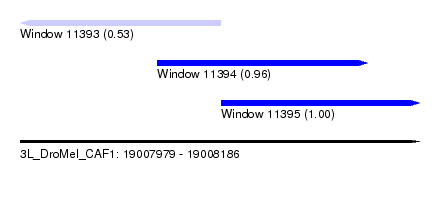
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,007,979 – 19,008,186 |
| Length | 207 |
| Max. P | 0.999039 |

| Location | 19,007,979 – 19,008,083 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.06 |
| Energy contribution | -22.94 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19007979 104 - 23771897 UCGAUGCCGCGAUGGAGGUGGAGUUGGCCCGUUGCUUAUGGAUGCACUGGGAAAAAAUUCCAUUGUUUUGUGACACUUUAAUCCUAA-GACAUUACAUUAAUAAU------- ..((((.(((((....(((((((((..((((.(((........))).))))....)))))))))...)))))...(((.......))-).))))...........------- ( -25.80) >DroSec_CAF1 141492 110 - 1 UCGAUGCCGCAAUGGAGGUGGAGUUGGCCCGUUGCUGAUGGAUGCACUGGGAA-AAAUUCCAUUGUUUUGUGACACUUUUAUCCUAA-GACAUUACAUUAAUAAUAGAUCAU ..(((..(((((....(((((((((..((((.(((........))).))))..-.)))))))))...)))))...(((.......))-)..................))).. ( -27.00) >DroSim_CAF1 147391 111 - 1 UCGAUGCCGCAAUGGAGGUGGAGUUGGCCCGUUGCUGAUGGAUGCACUGGGAAAAAAUUCCAUUGUUAUGUGACAGUUUAAUCCUAA-GACAUUACAUUAAUAAUGGAUCAU (((((.((((.......)))).)))))((((.(((........))).)))).......(((((((((((((((..((((.......)-))).))))).)))))))))).... ( -31.10) >DroEre_CAF1 143057 111 - 1 UCGAUGCCGCAAUGGAGGUGGAGUUGCCCCGUUGCUGAUGGAUGUACUGGGAA-AAAUUUCGCUGUCCUAGGACACUCAAAUCCCAAAAAAGUGACAUUAAUAAUGGGUCAA ..(((.(((((((((.(((......))))))))))(((((....((((((((.-...(((.(.((((....)))).).))))))).....)))).))))).....))))).. ( -30.20) >DroYak_CAF1 163303 104 - 1 UCGAUGCCGCGAUGGAGGUGGAGUUGGCCCGUUGCUGAUGGAUGUACUGGGAA-AAAUUUCACUGUCCUAGGACACUUAAAUCCUAG-AAAGUGGCAU------UGGGUCAU ((((((((((...(..(((((((((..((((.(((........))).))))..-.)))))))))..)((((((........))))))-...)))))))------)))..... ( -40.90) >consensus UCGAUGCCGCAAUGGAGGUGGAGUUGGCCCGUUGCUGAUGGAUGCACUGGGAA_AAAUUCCAUUGUUCUGUGACACUUUAAUCCUAA_GACAUUACAUUAAUAAUGGAUCAU .............((((((((((((..((((.(((........))).))))....)))))))))(((....))).......)))............................ (-24.06 = -22.94 + -1.12)

| Location | 19,008,050 – 19,008,159 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19008050 109 + 23771897 AACGGGC---CAACUCCACCUCCAUCGCGGCAUCGA-CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG ...(((.---....)))......((((.((((....-..))))..)))).(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... ( -32.90) >DroPse_CAF1 148996 87 + 1 --------------------------AUGGCUUUACCUUUACCUUUGCCUUUGCCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG --------------------------..(((...............))).(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... ( -26.96) >DroEre_CAF1 143135 109 + 1 AACGGGG---CAACUCCACCUCCAUUGCGGCAUCGA-CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG ...(((.---...).))((((...((((((((....-..))))...((.....)).)))).))))..(((((((.((.((((((((....)))))))))))))))))...... ( -36.00) >DroYak_CAF1 163374 109 + 1 AACGGGC---CAACUCCACCUCCAUCGCGGCAUCGA-CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG ...(((.---....)))......((((.((((....-..))))..)))).(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... ( -32.90) >DroAna_CAF1 139442 112 + 1 AAUGGGCAGCAAACUCCACCUCCGUCGUGGCAUCGA-CUUCGGUUCGAUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG ...............((((.......)))).(((((-(....).))))).(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... ( -32.90) >DroPer_CAF1 150135 87 + 1 --------------------------AUGGCUUUACCUUUACCUUUGCCUUUGCCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG --------------------------..(((...............))).(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... ( -26.96) >consensus AACGGGC___CAACUCCACCUCCAUCGCGGCAUCGA_CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCG ..................................................(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...... (-24.22 = -24.00 + -0.22)
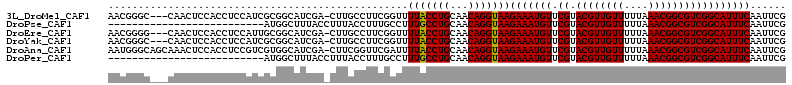


| Location | 19,008,083 – 19,008,186 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19008083 103 + 23771897 CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUGAUUUUCAUUUUUUGUACGACUCU----- .......(((...(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))..........................)))....----- ( -26.20) >DroPse_CAF1 149007 96 + 1 UUUACCUUUGCCUUUGCCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUUAUUUUCAUUUUU-------CUCU----- .((((((((((........)))).))))))(((((((.((.((((((((....)))))))))))))))))......................-------....----- ( -26.30) >DroSim_CAF1 147502 103 + 1 CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUGAUUUUCAUUUUUUGUACGACUCU----- .......(((...(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))..........................)))....----- ( -26.20) >DroEre_CAF1 143168 101 + 1 CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUGAUUUUCAUUUUUUGUCCGACU------- .......((((..(((((((...)))))))(((((((.((.((((((((....))))))))))))))))).........................))))..------- ( -28.40) >DroYak_CAF1 163407 103 + 1 CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUGAUUUUCAUUUUUUAUCCGACUCU----- .......((((..(((((((...)))))))(((((((.((.((((((((....))))))))))))))))).........................))))....----- ( -28.40) >DroPer_CAF1 150146 101 + 1 UUUACCUUUGCCUUUGCCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUUAUUUUCAUUUUU-------CUCUCUCUC .((((((((((........)))).))))))(((((((.((.((((((((....)))))))))))))))))......................-------......... ( -26.30) >consensus CUUGCCUUCGGUUUUACCUGCAACAGGUAAGAAAUGUUCGUACGUUGUUUUUAAACGGCGUCGGCAUUUCAAUUCGCUUGAUUUUCAUUUUUUGU_CGACUCU_____ .............(((((((...)))))))(((((((.((.((((((((....)))))))))))))))))...................................... (-24.22 = -24.00 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:50 2006