| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,000,091 – 19,000,188 |
| Length | 97 |
| Max. P | 0.665020 |

| Location | 19,000,091 – 19,000,188 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
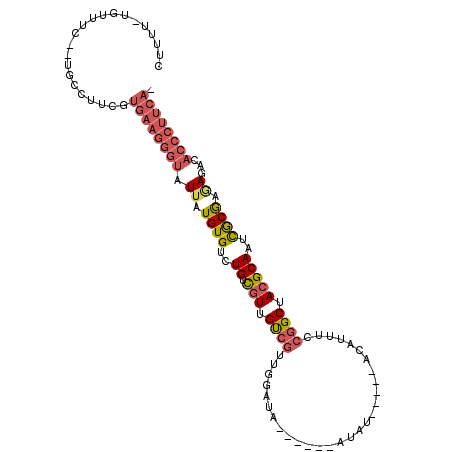
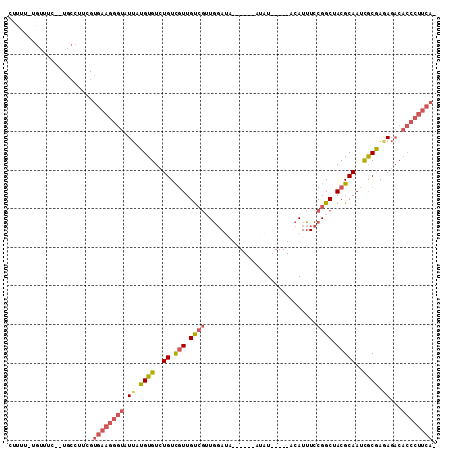
>3L_DroMel_CAF1 19000091 97 - 23771897 CUUUUGUGUUUC--UGCCAUCGUGAAGGGUAUUAUGUGUCUGUCGUUGUCAUUGGAUA------AUAU-----ACAUUUCCGGCUACGCAAUCGCGAGAGACACCCUUCA- .....(((((((--((((..(.....)((....((((((.....((((((....))))------))))-----))))..)))))..(((....)))))))))))......- ( -24.70) >DroSec_CAF1 133678 97 - 1 CUUUUGUGUUUC--UGCCUUCGUGAAGGGUAUUAUGUGUCUGUCGUUGCCGAUGGAUA------AUAU-----ACAUUUCCGGCUACGCAACCGCGAGAGACACCCUUCA- .....(((((((--((((...(.((((.((((....(((((((((....)))))))))------..))-----)).))))))))..(((....)))))))))))......- ( -30.20) >DroSim_CAF1 139028 97 - 1 CUUUUGUGUUUC--UGCCUUCGUGAAGGGUAUUAUGUGUCUGUCGUUGUCGUUGGAUA------AUAU-----ACAUUUCCGGCAACGCAAUCGCGAGAGACACCCUUCA- .....(((((((--((((((.....)))))....((((..((.((((((((.((....------....-----.))....))))))))))..))))))))))))......- ( -28.60) >DroEre_CAF1 134962 93 - 1 CUUUU-CGUUUC--UGCCUCUGUGAAGGGUAUUAUGUGUCUGUCGUUGUCGUUGGA---------UAU-----ACAUUUCCGGCUACGCAAUCGCAAAAGACACCCUUCA- .....-......--........((((((((.((.((((..((.(((....((((((---------...-----.....)))))).)))))..)))).))...))))))))- ( -25.80) >DroYak_CAF1 155181 94 - 1 CUUUU-UGUUUC--UGCCACCGUGAAGGGUAUUAUGUGUCUGUCGUUGUCGUUGGA---------UAU-----ACAUUUCCGGCUACGCAAUUGCAAAAGACACCCUUCAU .....-......--.......(((((((((.......((((((.(((((.((((((---------...-----.....))))))...))))).))...))))))))))))) ( -25.31) >DroAna_CAF1 131365 99 - 1 UGCUU-UGUUUUCAUUGAGGUGUUGAGGGUAUUGUGUGUCUGUUUUUGUGGAUAUAUAUAGUAAGUAUACCCGACAUUUCCGGCUACGCAAUUACGACAU----------- (((..-..........((((((((..((((((((((((((..(....)..))))))))........)))))))))))))).(....))))..........----------- ( -22.90) >consensus CUUUU_UGUUUC__UGCCUUCGUGAAGGGUAUUAUGUGUCUGUCGUUGUCGUUGGAUA______AUAU_____ACAUUUCCGGCUACGCAAUCGCGAGAGACACCCUUCA_ ......................((((((((.((.((((..((.(((.((((.............................)))).)))))..)))).))...)))))))). (-15.86 = -16.83 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:46 2006