| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,991,537 – 18,991,644 |
| Length | 107 |
| Max. P | 0.953964 |

| Location | 18,991,537 – 18,991,644 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
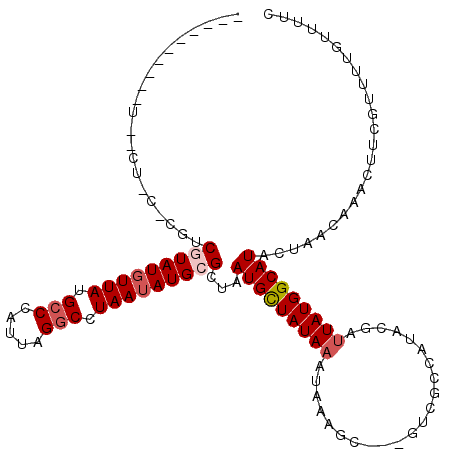
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
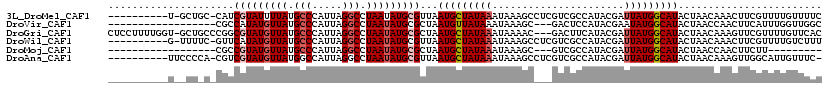
>3L_DroMel_CAF1 18991537 107 + 23771897 ----------U-GCUGC-CAUCGUAUUUUAUGCCCAUUAGGCCUAAUAUGCGUUAAUGCUAUAAAUAAAGCCUCGUCGCCAUACGAUUAUGGCAUACUAACAAACUUCGUUUUGUUUUC ----------.-..(((-((((((((.(((.(((.....))).)))...(((.....(((........))).....))).))))))...)))))....((((((......))))))... ( -21.90) >DroVir_CAF1 140468 98 + 1 ------------------CGCCAUAUGUUAUGCCCAUUAGGCCUAAUAUGCGCUAAUGUUAUAAAUAAAGC---GACUCCAUACGAAUAUGGCAUACUAACCAACUUCAUUUGGUUGGC ------------------.(((((((((((.(((.....))).))))...((((..............)))---)...........)))))))...((((((((......)))))))). ( -26.24) >DroGri_CAF1 127983 115 + 1 CUCCUUUUGGU-GCUGCCCGGCGUAUGUUAUGCCCAUUAGGCCUAAUAUGCGCUAAUGCUAUAAAUAAAAC---GACUUCAUACGAUUAUGGCAUACUAACAAAGUUCGUUUUGUUCAC .......((((-(.((((.(((((((((((.(((.....))).)))))))))))................(---(........)).....)))))))))((((((....)))))).... ( -28.90) >DroWil_CAF1 162361 107 + 1 ----------G-UUUUC-GUUCAUAUGUUAUGCCCAUUAGGCCUAAUAUGCGUUAAUGCUAUAAAUAAAGCCUCGUCGCCAUACGAUUAUGGCAUACUAACAAACUUCGUUUUGUCUUU ----------(-(((..-.....(((((((.(((.....))).))))))).(((((((((((((........((((......)))))))))))))..)))))))).............. ( -20.00) >DroMoj_CAF1 145401 89 + 1 ------------------CGCCGUAUGUUAUGCCCAUUAGGCCUAAUAUGCGCUAAUGCUAUAAAUAAAGC---GUCGCCAUACGAUUAUGGCAUACUAACCAACUUCUU--------- ------------------.((.((((((((.(((.....))).))))))))))..(((((........)))---)).((((((....)))))).................--------- ( -24.60) >DroAna_CAF1 123062 107 + 1 ----------UUCCCCA-CGUCGUAUGUUAUGGCCAUUAGGCCUAAUAUGCGUUAAUGCUAUAAAUAAAGCCUCGUCGCCAUACGAUUAUGGCAUACUAACAAAGUUGGCAUUGUUUC- ----------.......-...(((((((((.((((....))))))))))))).((((((((........(((..((((.....))))...)))..(((.....)))))))))))....- ( -31.10) >consensus __________U__CU_C_CGUCGUAUGUUAUGCCCAUUAGGCCUAAUAUGCGCUAAUGCUAUAAAUAAAGC___GUCGCCAUACGAUUAUGGCAUACUAACAAACUUCGUUUUGUUUUC .....................(((((((((.(((.....))).)))))))))...(((((((((......................)))))))))........................ (-15.27 = -15.97 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:44 2006