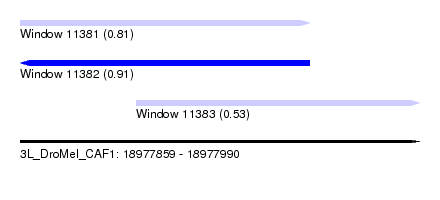
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,977,859 – 18,977,990 |
| Length | 131 |
| Max. P | 0.908748 |

| Location | 18,977,859 – 18,977,954 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.36 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18977859 95 + 23771897 CAUUUUUCAUUUGCGCGAGCCGCGUCCUUGCCAAG--GACCCCGCCCAUUUG-----------AGUAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA .....((((..((.(((.(..(.(((((.....))--)))))))).))..))-----------)).((((((........))))))...((((((((...)))))))) ( -24.30) >DroPse_CAF1 118386 83 + 1 CAUUUCUCAUUUG--AGAGCUGCGUCCUUGCCGGGCAGGCCCCGCCCAG-----------------------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA ..........(((--((((((..(((((.((.((((.(....))))).)-----------------------))))))..)))))))))((((((((...)))))))) ( -34.90) >DroEre_CAF1 112966 88 + 1 -------CAUUUGUGCGAGCCGCGUCCUUGCCAAG--GACCCCGCCCAUUUG-----------AGUAGAGACCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA -------.........((((((.((((((((((((--(.......))..)))-----------.)))......))))).))))))....((((((((...)))))))) ( -22.00) >DroYak_CAF1 128149 94 + 1 CAUUUUUCAUUUGUGCAAGCCGCGUCCUUGCCAAG--GACCCCGCCCAUUUG-----------AGUAGAG-CCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA .........((..(((((((.(.(((((.....))--))).).)).......-----------...((((-(((.....)))))))...)))))..)).......... ( -24.50) >DroAna_CAF1 109752 106 + 1 CAUUUUUCAUUUGUGCGAGUCGCGUCCUUGCCAAG--GACCCCGCCCAUUUUCAGUGUUUUCCAGCUCAGCCGAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA ................((((((.(((((((....(--((...(((.........)))...))).((...))))))))).))))))....((((((((...)))))))) ( -24.00) >DroPer_CAF1 119699 83 + 1 CAUUUCUCAUUUG--AGAGCUGCGUCCUUGCCGGGCAGGCCCCGCCCAG-----------------------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA ..........(((--((((((..(((((.((.((((.(....))))).)-----------------------))))))..)))))))))((((((((...)))))))) ( -34.90) >consensus CAUUUUUCAUUUGUGCGAGCCGCGUCCUUGCCAAG__GACCCCGCCCAUUUG___________AGUAGAG_CCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAA ................((((((.(((((.............................................))))).))))))....((((((((...)))))))) (-16.86 = -16.36 + -0.50)


| Location | 18,977,859 – 18,977,954 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18977859 95 - 23771897 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGGCUCUACU-----------CAAAUGGGCGGGGUC--CUUGGCAAGGACGCGGCUCGCGCAAAUGAAAAAUG ((((....(((....)))......((((.((((((((((((..((-----------(....)))..)))))--)......)))))).))))...)))).......... ( -30.90) >DroPse_CAF1 118386 83 - 1 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUG-----------------------CUGGGCGGGGCCUGCCCGGCAAGGACGCAGCUCU--CAAAUGAGAAAUG ((((((.......))))))......((..(((((((-----------------------((((((((...))))))))).))))))..))(((--(....)))).... ( -35.10) >DroEre_CAF1 112966 88 - 1 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGUCUCUACU-----------CAAAUGGGCGGGGUC--CUUGGCAAGGACGCGGCUCGCACAAAUG------- ((((((.......)))))).....((((.((((((((.(((..((-----------(....)))..))).)--)......)))))).))))..........------- ( -27.20) >DroYak_CAF1 128149 94 - 1 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGG-CUCUACU-----------CAAAUGGGCGGGGUC--CUUGGCAAGGACGCGGCUUGCACAAAUGAAAAAUG ((((((.......))))))....(((((.((((((((-(((..((-----------(....)))..)))))--.......)))))).)))))................ ( -28.51) >DroAna_CAF1 109752 106 - 1 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUCGGCUGAGCUGGAAAACACUGAAAAUGGGCGGGGUC--CUUGGCAAGGACGCGACUCGCACAAAUGAAAAAUG ((((((.......))))))......((((.(.(((((.(((((.((.....)))).....))).))))).)--..))))......(((...))).............. ( -23.10) >DroPer_CAF1 119699 83 - 1 UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUG-----------------------CUGGGCGGGGCCUGCCCGGCAAGGACGCAGCUCU--CAAAUGAGAAAUG ((((((.......))))))......((..(((((((-----------------------((((((((...))))))))).))))))..))(((--(....)))).... ( -35.10) >consensus UUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGG_CUCUACU___________CAAAUGGGCGGGGUC__CUUGGCAAGGACGCGGCUCGCACAAAUGAAAAAUG ((((((.......)))))).....((((.((((((............................((.(((....))).)).)))))).))))................. (-19.43 = -19.48 + 0.06)

| Location | 18,977,897 – 18,977,990 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18977897 93 + 23771897 CCCGCCCAUUUG-----------AGUAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUG---- ............-----------.(((((.(...((((...(((((...((((((((...))))))))..))))).))))...))))))...............---- ( -23.00) >DroPse_CAF1 118424 85 + 1 CCCGCCCAG-----------------------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUAAUUUUCCCU ........(-----------------------((((((...(((((...((((((((...))))))))..))))).))))((((.......))))))).......... ( -18.40) >DroSim_CAF1 116730 93 + 1 CCCGCCCAUUUA-----------UGCAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUG---- ..........((-----------(((((((....((((...(((((...((((((((...))))))))..))))).))))...(....).))))))))).....---- ( -22.30) >DroEre_CAF1 112997 93 + 1 CCCGCCCAUUUG-----------AGUAGAGACCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCACAGCAGUCCCAAGUCUACAUUUUGUAUACUUUG---- ...........(-----------((((.((((..((((...(((.....((((((((...))))))))....))).))))...))))((.....)).)))))..---- ( -20.80) >DroAna_CAF1 109790 104 + 1 CCCGCCCAUUUUCAGUGUUUUCCAGCUCAGCCGAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUCCGUUUUGUAUACUUUG---- .............(((((.....(((..((.(..((((...(((((...((((((((...))))))))..))))).))))...).))..)))....)))))...---- ( -21.20) >DroPer_CAF1 119737 85 + 1 CCCGCCCAG-----------------------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUAAUUUUCCCU ........(-----------------------((((((...(((((...((((((((...))))))))..))))).))))((((.......))))))).......... ( -18.40) >consensus CCCGCCCAUUU____________AG_AGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUG____ ..................................((((...(((((...((((((((...))))))))..))))).))))............................ (-17.23 = -17.40 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:39 2006